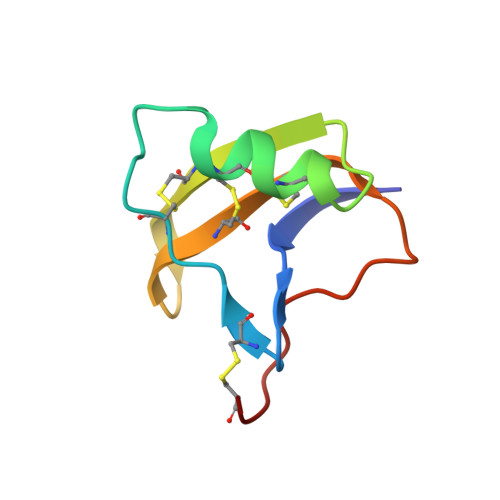
Refined structure of BeM9 reveals arginine hand, an overlooked structural motif in scorpion toxins affecting sodium channels.
Kuldyushev, N.A., Mineev, K.S., Berkut, A.A., Peigneur, S., Arseniev, A.S., Tytgat, J., Grishin, E.V., Vassilevski, A.A.(2018) Proteins 86: 1117-1122
- PubMed: 30007037
- DOI: https://doi.org/10.1002/prot.25583
- Primary Citation of Related Structures:
5MOU - PubMed Abstract:
Sodium channel alpha-toxins from scorpion venom (α-NaTx) inhibit the inactivation of voltage-gated sodium channels. We used solution NMR to investigate the structure of BeM9 toxin from Mesobuthus eupeus scorpion, a prototype α-NaTx classified as an "α-like" toxin due to its wide spectrum of activity on insect and mammalian channels. We identified a new motif that we named "arginine hand," whereby arginine side chain forms several hydrogen bonds with main chain atoms. The arginine hand was found in the "specificity module," a part of the molecule that dictates toxin selectivity; and just single arginine-to-lysine point mutation drastically changed BeM9 selectivity profile.
- Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russia.
Organizational Affiliation: