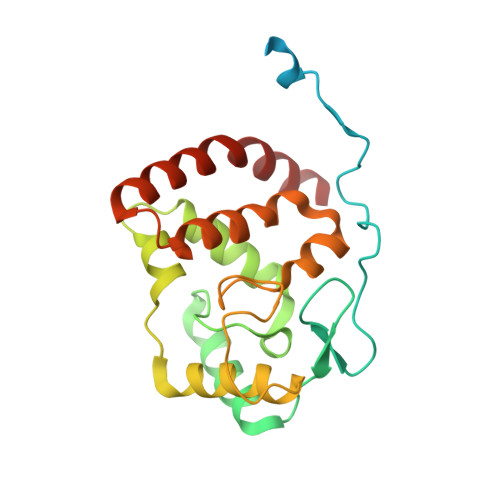
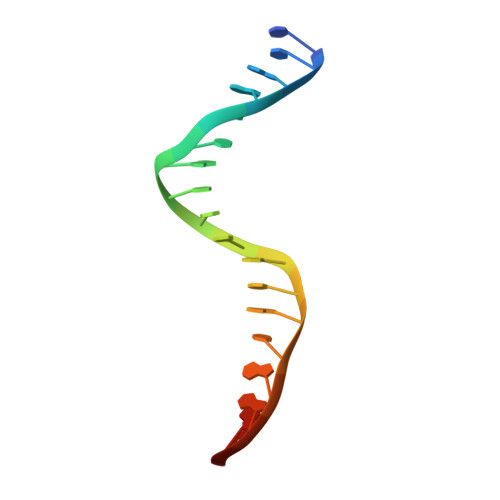
The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Tunnicliffe, R.B., Lockhart-Cairns, M.P., Levy, C., Mould, A.P., Jowitt, T.A., Sito, H., Baldock, C., Sandri-Goldin, R.M., Golovanov, A.P.(2017) Nucleic Acids Res 45: 8064-8078
- PubMed: 28505309
- DOI: https://doi.org/10.1093/nar/gkx419
- Primary Citation of Related Structures:
5MHJ, 5MHK - PubMed Abstract:
The transcription factor ICP4 from herpes simplex virus has a central role in regulating the gene expression cascade which controls viral infection. Here we present the crystal structure of the functionally essential ICP4 DNA binding domain in complex with a segment from its own promoter, revealing a novel homo-dimeric fold. We also studied the complex in solution by small angle X-Ray scattering, nuclear magnetic resonance and surface-plasmon resonance which indicated that, in addition to the globular domain, a flanking intrinsically disordered region also recognizes DNA. Together the data provides a rationale for the bi-partite nature of the ICP4 DNA recognition consensus sequence as the globular and disordered regions bind synergistically to adjacent DNA motifs. Therefore in common with its eukaryotic host, the viral transcription factor ICP4 utilizes disordered regions to enhance the affinity and tune the specificity of DNA interactions in tandem with a globular domain.
- Manchester Institute of Biotechnology, School of Chemistry, Faculty of Science and Engineering, The University of Manchester, Manchester M1 7DN, UK.
Organizational Affiliation: