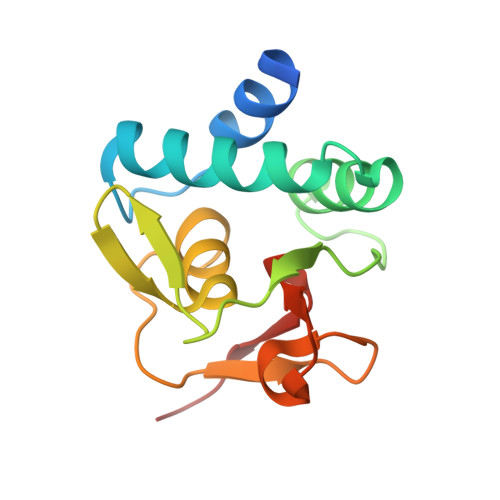
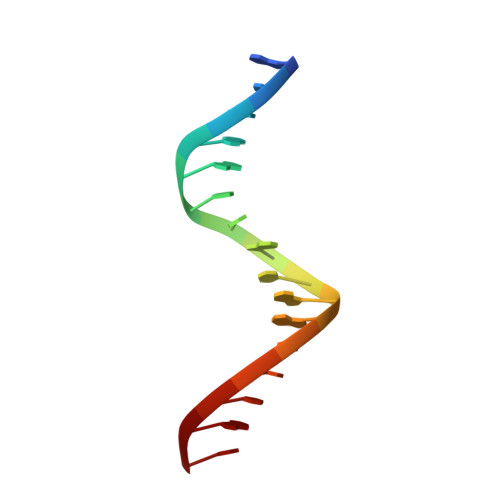
Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Martin-Malpartida, P., Batet, M., Kaczmarska, Z., Freier, R., Gomes, T., Aragon, E., Zou, Y., Wang, Q., Xi, Q., Ruiz, L., Vea, A., Marquez, J.A., Massague, J., Macias, M.J.(2017) Nat Commun 8: 2070-2070
- PubMed: 29234012
- DOI: https://doi.org/10.1038/s41467-017-02054-6
- Primary Citation of Related Structures:
5MEY, 5MEZ, 5MF0, 5NM9, 5OD6, 5ODG - PubMed Abstract:
Smad transcription factors activated by TGF-β or by BMP receptors form trimeric complexes with Smad4 to target specific genes for cell fate regulation. The CAGAC motif has been considered as the main binding element for Smad2/3/4, whereas Smad1/5/8 have been thought to preferentially bind GC-rich elements. However, chromatin immunoprecipitation analysis in embryonic stem cells showed extensive binding of Smad2/3/4 to GC-rich cis-regulatory elements. Here, we present the structural basis for specific binding of Smad3 and Smad4 to GC-rich motifs in the goosecoid promoter, a nodal-regulated differentiation gene. The structures revealed a 5-bp consensus sequence GGC(GC)|(CG) as the binding site for both TGF-β and BMP-activated Smads and for Smad4. These 5GC motifs are highly represented as clusters in Smad-bound regions genome-wide. Our results provide a basis for understanding the functional adaptability of Smads in different cellular contexts, and their dependence on lineage-determining transcription factors to target specific genes in TGF-β and BMP pathways.
- Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Baldiri Reixac, 10, 08028, Barcelona, Spain.
Organizational Affiliation: