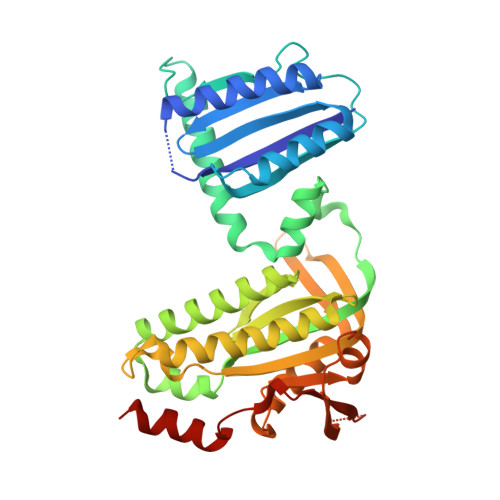
Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
Lindner, R., Hartmann, E., Tarnawski, M., Winkler, A., Frey, D., Reinstein, J., Meinhart, A., Schlichting, I.(2017) J Mol Biology 429: 1336-1351
- PubMed: 28336405
- DOI: https://doi.org/10.1016/j.jmb.2017.03.020
- Primary Citation of Related Structures:
5M27, 5M2A, 5MBB, 5MBC, 5MBD, 5MBE, 5MBG, 5MBH, 5MBJ, 5MBK, 5NBY - PubMed Abstract:
Light-regulated enzymes enable organisms to quickly respond to changing light conditions. We characterize a photoactivatable adenylyl cyclase (AC) from Beggiatoa sp. (bPAC) that translates a blue light signal into the production of the second messenger cyclic AMP. bPAC contains a BLUF photoreceptor domain that senses blue light using a flavin chromophore, linked to an AC domain. We present a dark state crystal structure of bPAC that closely resembles the recently published structure of the homologous OaPAC from Oscillatoria acuminata. To elucidate the structural mechanism of light-dependent AC activation by the BLUF domain, we determined the crystal structures of illuminated bPAC and of a pseudo-lit state variant. We use hydrogen-deuterium exchange measurements of secondary structure dynamics and hypothesis-driven point mutations to trace the activation pathway from the chromophore in the BLUF domain to the active site of the cyclase. The structural changes are relayed from the residues interacting with the excited chromophore through a conserved kink of the BLUF β-sheet to a tongue-like extrusion of the AC domain that regulates active site opening and repositions catalytic residues. Our findings not only show the specific molecular pathway of photoactivation in BLUF-regulated ACs but also have implications for the general understanding of signaling in BLUF domains and of the activation of ACs.
- Max Planck Institute for Medical Research, Jahnstr, 29, 69120 Heidelberg, Germany.
Organizational Affiliation: