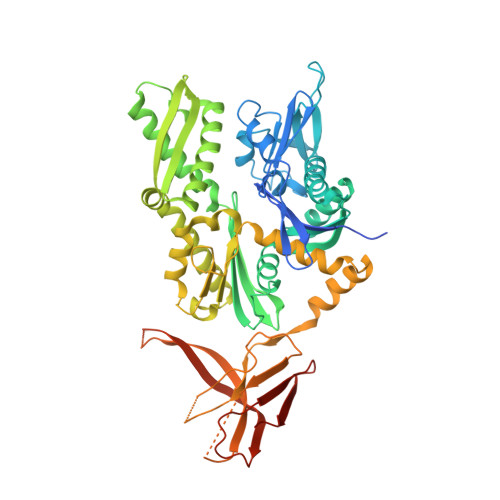
Structural insights into a unique Hsp70-Hsp40 interaction in the eukaryotic ribosome-associated complex.
Weyer, F.A., Gumiero, A., Gese, G.V., Lapouge, K., Sinning, I.(2017) Nat Struct Mol Biol 24: 144-151
- PubMed: 28067917
- DOI: https://doi.org/10.1038/nsmb.3349
- Primary Citation of Related Structures:
5MB9 - PubMed Abstract:
Cotranslational chaperones assist de novo folding of nascent polypeptides, prevent them from aggregating and modulate translation. The ribosome-associated complex (RAC) is unique in that the Hsp40 protein Zuo1 and the atypical Hsp70 chaperone Ssz1 form a stable heterodimer, which acts as a cochaperone for the Hsp70 chaperone Ssb. Here we present the structure of the Chaetomium thermophilum RAC core comprising Ssz1 and the Zuo1 N terminus. We show how the conserved allostery of Hsp70 proteins is abolished and this Hsp70-Hsp40 pair is molded into a functional unit. Zuo1 stabilizes Ssz1 in trans through interactions that in canonical Hsp70s occur in cis. Ssz1 is catalytically inert and cannot adopt the closed conformation, but the substrate binding domain β is completed by Zuo1. Our study offers insights into the coupling of a special Hsp70-Hsp40 pair, which evolved to link protein folding and translation.
- Heidelberg University Biochemistry Center (BZH), Heidelberg, Germany.
Organizational Affiliation: