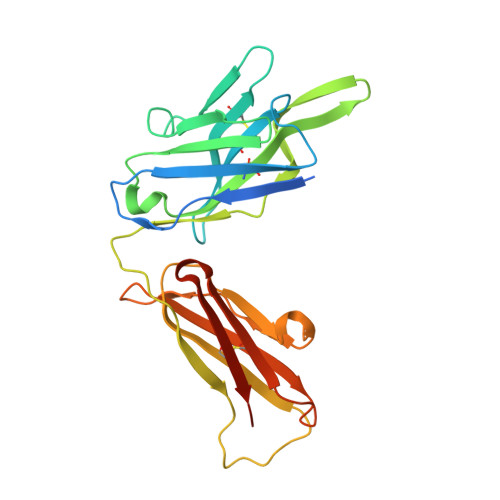
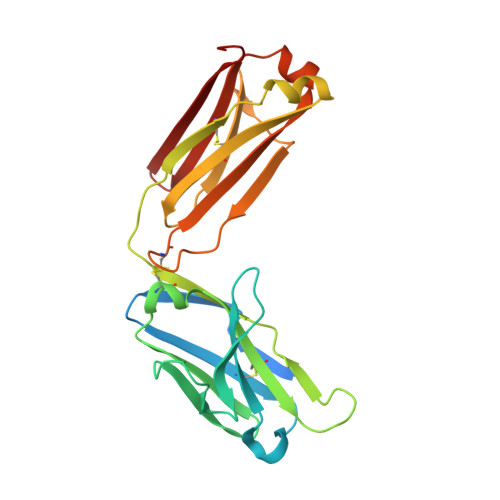
Structure of a protective epitope of group B Streptococcus type III capsular polysaccharide.
Carboni, F., Adamo, R., Fabbrini, M., De Ricco, R., Cattaneo, V., Brogioni, B., Veggi, D., Pinto, V., Passalacqua, I., Oldrini, D., Rappuoli, R., Malito, E., Margarit, I.Y.R., Berti, F.(2017) Proc Natl Acad Sci U S A 114: 5017-5022
- PubMed: 28439022
- DOI: https://doi.org/10.1073/pnas.1701885114
- Primary Citation of Related Structures:
5M63 - PubMed Abstract:
Despite substantial progress in the prevention of group B Streptococcus (GBS) disease with the introduction of intrapartum antibiotic prophylaxis, this pathogen remains a leading cause of neonatal infection. Capsular polysaccharide conjugate vaccines have been tested in phase I/II clinical studies, showing promise for further development. Mapping of epitopes recognized by protective antibodies is crucial for understanding the mechanism of action of vaccines and for enabling antigen design. In this study, we report the structure of the epitope recognized by a monoclonal antibody with opsonophagocytic activity and representative of the protective response against type III GBS polysaccharide. The structure and the atomic-level interactions were determined by saturation transfer difference (STD)-NMR and X-ray crystallography using oligosaccharides obtained by synthetic and depolymerization procedures. The GBS PSIII epitope is made by six sugars. Four of them derive from two adjacent repeating units of the PSIII backbone and two of them from the branched galactose-sialic acid disaccharide contained in this sequence. The sialic acid residue establishes direct binding interactions with the functional antibody. The crystal structure provides insight into the molecular basis of antibody-carbohydrate interactions and confirms that the conformational epitope is not required for antigen recognition. Understanding the structural basis of immune recognition of capsular polysaccharide epitopes can aid in the design of novel glycoconjugate vaccines.
- GSK Vaccines, 53100 Siena, Italy.
Organizational Affiliation: