Comment on "Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2".
Zhang, Y., Justin, N., Wilson, J.R., Gamblin, S.J.(2016) Science 354: 1543-1543
- PubMed: 28008037
- DOI: https://doi.org/10.1126/science.aaf6236
- Primary Citation of Related Structures:
5M5G - PubMed Abstract:
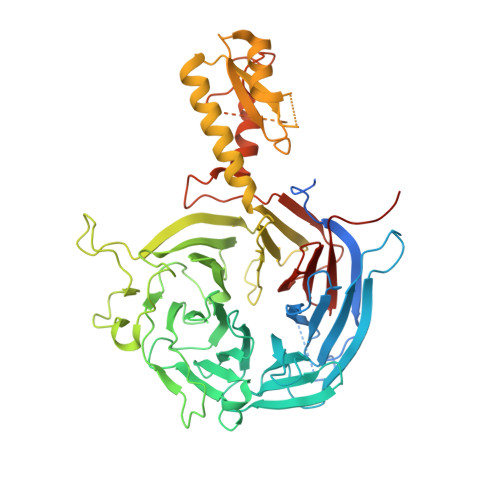
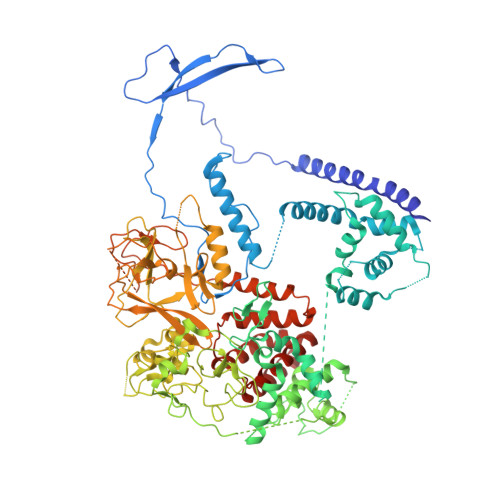
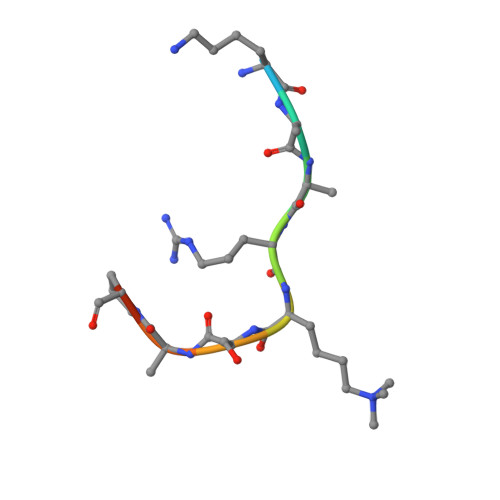
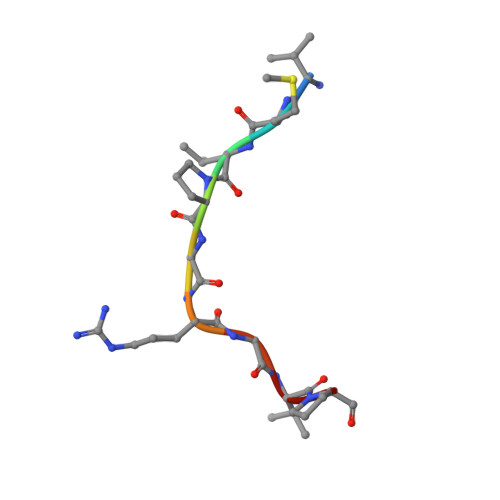
Jiao and Liu (Research Articles, 16 October 2015, aac4383) reported the crystal structure of the protein complex polycomb repressive complex 2 from Chaetomium thermophilum This landmark structure has brought invaluable insights into the activation mechanism of this essential methyltransferase. However, the analysis of the x-ray data discussed below suggests that the description of oncogenic H3K27M peptide binding to the active site is incorrect.
- Francis Crick Institute, 1 Midland Road, London, NW1 1AT, UK.
Organizational Affiliation: