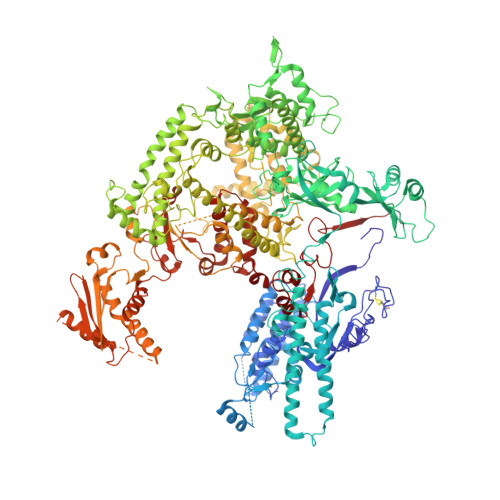
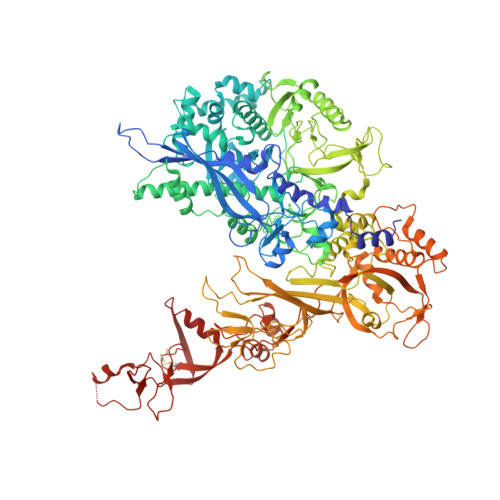
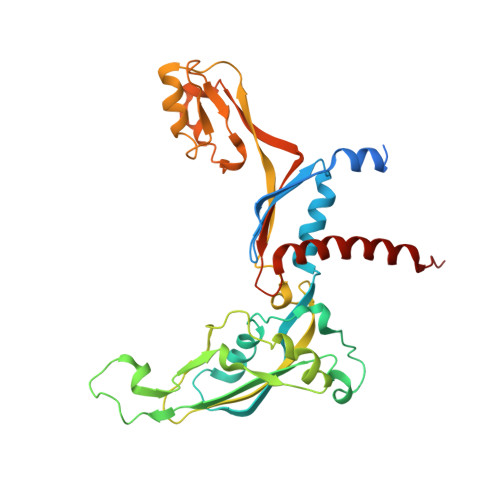
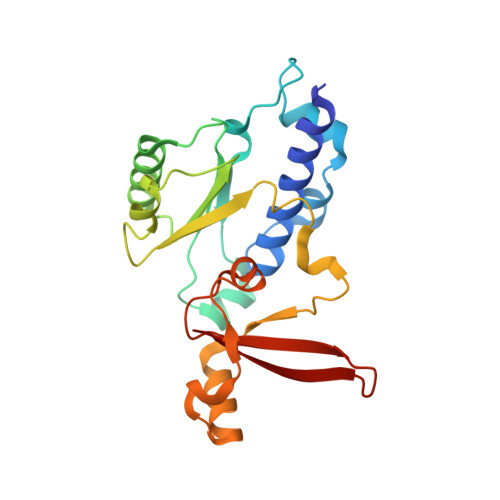
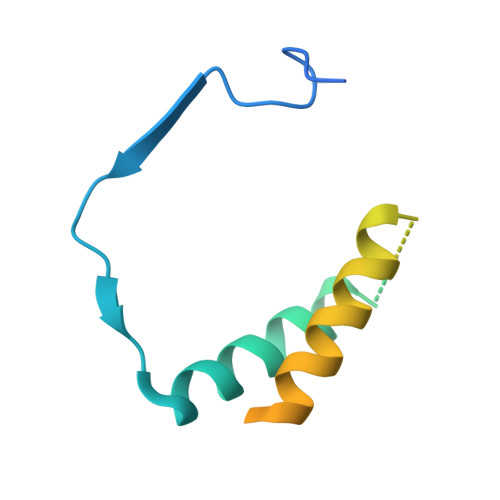
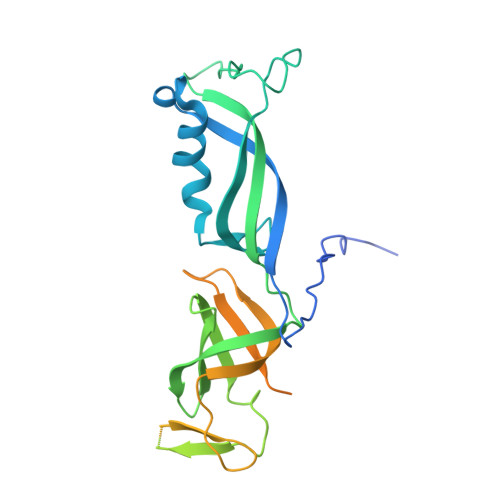
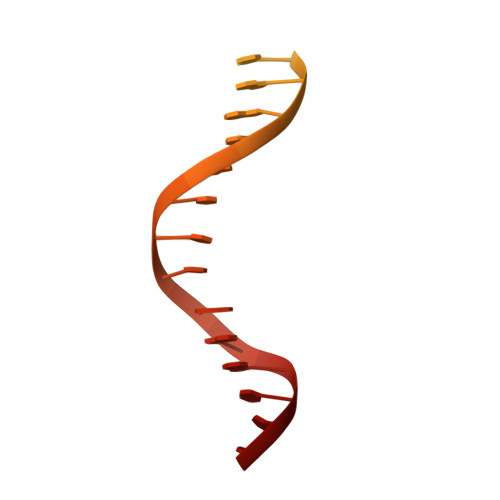
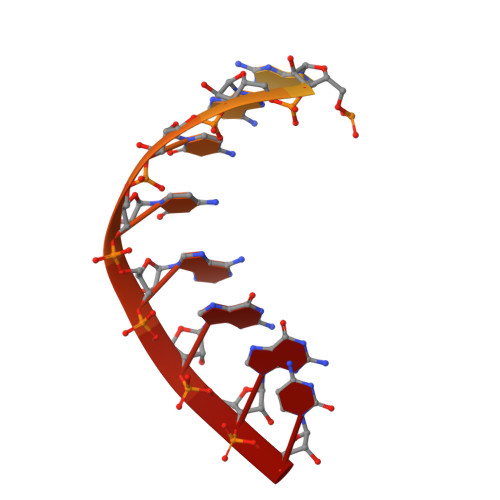
Structure of RNA polymerase I transcribing ribosomal DNA genes.
Neyer, S., Kunz, M., Geiss, C., Hantsche, M., Hodirnau, V.V., Seybert, A., Engel, C., Scheffer, M.P., Cramer, P., Frangakis, A.S.(2016) Nature 540: 607-610
- PubMed: 27842382
- DOI: https://doi.org/10.1038/nature20561
- Primary Citation of Related Structures:
5M3F, 5M3M - PubMed Abstract:
RNA polymerase I (Pol I) is a highly processive enzyme that transcribes ribosomal DNA (rDNA) and regulates growth of eukaryotic cells. Crystal structures of free Pol I from the yeast Saccharomyces cerevisiae have revealed dimers of the enzyme stabilized by a 'connector' element and an expanded cleft containing the active centre in an inactive conformation. The central bridge helix was unfolded and a Pol-I-specific 'expander' element occupied the DNA-template-binding site. The structure of Pol I in its active transcribing conformation has yet to be determined, whereas structures of Pol II and Pol III have been solved with bound DNA template and RNA transcript. Here we report structures of active transcribing Pol I from yeast solved by two different cryo-electron microscopy approaches. A single-particle structure at 3.8 Å resolution reveals a contracted active centre cleft with bound DNA and RNA, and a narrowed pore beneath the active site that no longer holds the RNA-cleavage-stimulating domain of subunit A12.2. A structure at 29 Å resolution that was determined from cryo-electron tomograms of Pol I enzymes transcribing cellular rDNA confirms contraction of the cleft and reveals that incoming and exiting rDNA enclose an angle of around 150°. The structures suggest a model for the regulation of transcription elongation in which contracted and expanded polymerase conformations are associated with active and inactive states, respectively.
- Max-Planck-Institute for Biophysical Chemistry, Department of Molecular Biology, Am Fassberg 11, 37077 Göttingen, Germany.
Organizational Affiliation: