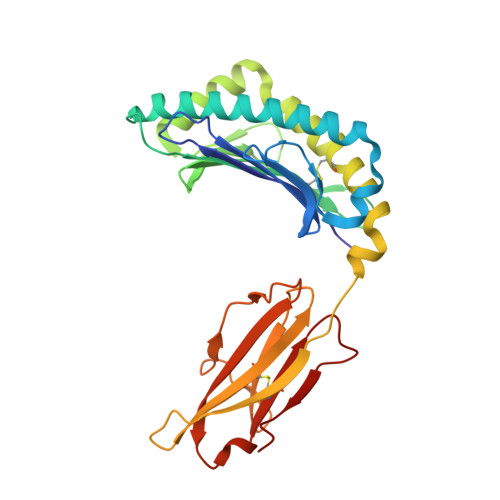
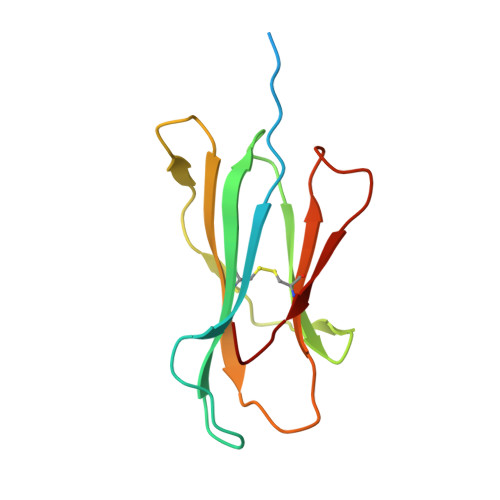
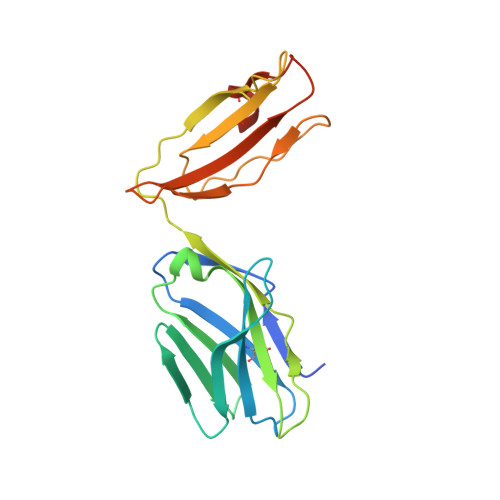
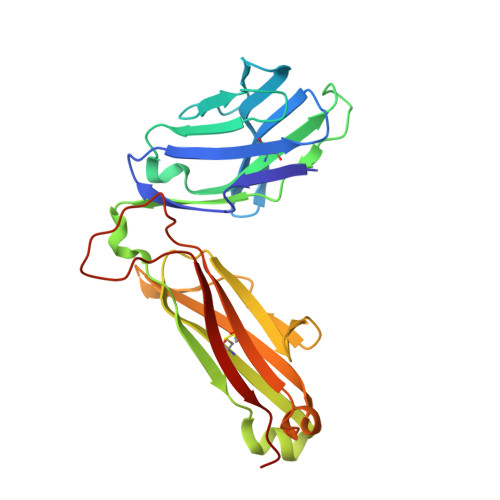
Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
Allerbring, E., Duru, A., Sun, R., Han, X., Uchtenhagen, H., Madhurantakam, C., Popov, A., Markova, N., Talyzina, A., Nygren, P.A., Sandalova, T., Achour, A.To be published.