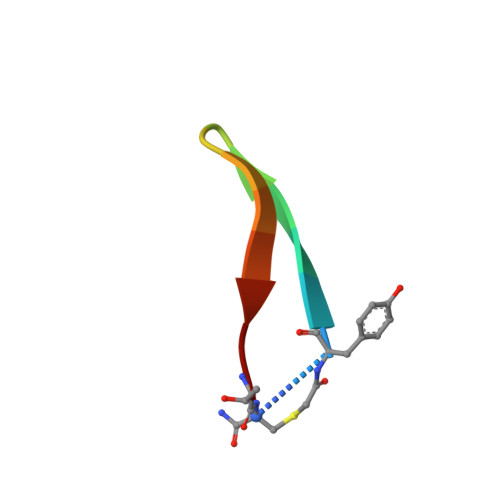
Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Kawamura, A., Munzel, M., Kojima, T., Yapp, C., Bhushan, B., Goto, Y., Tumber, A., Katoh, T., King, O.N., Passioura, T., Walport, L.J., Hatch, S.B., Madden, S., Muller, S., Brennan, P.E., Chowdhury, R., Hopkinson, R.J., Suga, H., Schofield, C.J.(2017) Nat Commun 8: 14773-14773
- PubMed: 28382930
- DOI: https://doi.org/10.1038/ncomms14773
- Primary Citation of Related Structures:
5LY1, 5LY2 - PubMed Abstract:
The JmjC histone demethylases (KDMs) are linked to tumour cell proliferation and are current cancer targets; however, very few highly selective inhibitors for these are available. Here we report cyclic peptide inhibitors of the KDM4A-C with selectivity over other KDMs/2OG oxygenases, including closely related KDM4D/E isoforms. Crystal structures and biochemical analyses of one of the inhibitors (CP2) with KDM4A reveals that CP2 binds differently to, but competes with, histone substrates in the active site. Substitution of the active site binding arginine of CP2 to N-ɛ-trimethyl-lysine or methylated arginine results in cyclic peptide substrates, indicating that KDM4s may act on non-histone substrates. Targeted modifications to CP2 based on crystallographic and mass spectrometry analyses results in variants with greater proteolytic robustness. Peptide dosing in cells manifests KDM4A target stabilization. Although further development is required to optimize cellular activity, the results reveal the feasibility of highly selective non-metal chelating, substrate-competitive inhibitors of the JmjC KDMs.
- Chemistry Research Laboratory, Department of Chemistry, University of Oxford, 12 Mansfield Road, Oxford OX1 3TA, UK.
Organizational Affiliation: