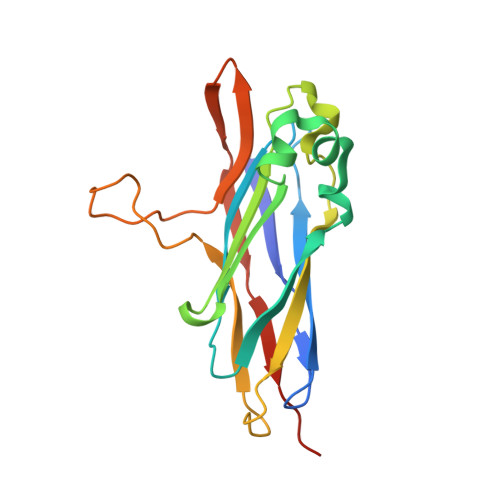
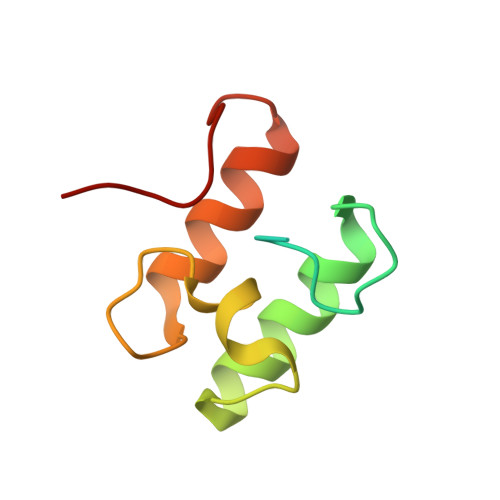
Single Binding Mode Integration of Hemicellulose-degrading Enzymes via Adaptor Scaffoldins in Ruminococcus flavefaciens Cellulosome.
Bule, P., Alves, V.D., Leitao, A., Ferreira, L.M., Bayer, E.A., Smith, S.P., Gilbert, H.J., Najmudin, S., Fontes, C.M.(2016) J Biological Chem 291: 26658-26669
- PubMed: 27875311
- DOI: https://doi.org/10.1074/jbc.M116.761643
- Primary Citation of Related Structures:
5LXV - PubMed Abstract:
The assembly of one of Nature's most elaborate multienzyme complexes, the cellulosome, results from the binding of enzyme-borne dockerins to reiterated cohesin domains located in a non-catalytic primary scaffoldin. Generally, dockerins present two similar cohesin-binding interfaces that support a dual binding mode. The dynamic integration of enzymes in cellulosomes, afforded by the dual binding mode, is believed to incorporate additional flexibility in highly populated multienzyme complexes. Ruminococcus flavefaciens, the primary degrader of plant structural carbohydrates in the rumen of mammals, uses a portfolio of more than 220 different dockerins to assemble the most intricate cellulosome known to date. A sequence-based analysis organized R. flavefaciens dockerins into six groups. Strikingly, a subset of R. flavefaciens cellulosomal enzymes, comprising dockerins of groups 3 and 6, were shown to be indirectly incorporated into primary scaffoldins via an adaptor scaffoldin termed ScaC. Here, we report the crystal structure of a group 3 R. flavefaciens dockerin, Doc3, in complex with ScaC cohesin. Doc3 is unusual as it presents a large cohesin-interacting surface that lacks the structural symmetry required to support a dual binding mode. In addition, dockerins of groups 3 and 6, which bind exclusively to ScaC cohesin, display a conserved mechanism of protein recognition that is similar to Doc3. Groups 3 and 6 dockerins are predominantly appended to hemicellulose-degrading enzymes. Thus, single binding mode dockerins interacting with adaptor scaffoldins exemplify an evolutionary pathway developed by R. flavefaciens to recruit hemicellulases to the sophisticated cellulosomes acting in the gastrointestinal tract of mammals.
- From the CIISA-Faculdade de Medicina Veterinária, ULisboa, Pólo Universitário do Alto da Ajuda, Avenida da Universidade Técnica, 1300-477 Lisboa, Portugal.
Organizational Affiliation: