Structure of the DNA-binding domain of LUX ARRHYTHMO
Zubieta, C., Silva, C.S., Lai, X., Wigge, P., Nanao, M.H., Nayak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription factor LUX | 57 | Arabidopsis thaliana | Mutation(s): 0 Gene Names: LUX, PCL1, At3g46640, F12A12.160 | 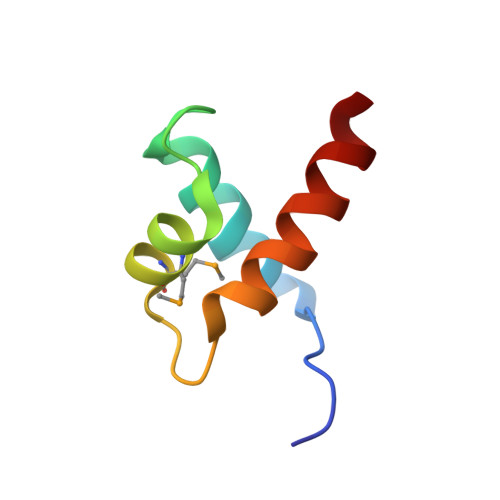 | |
UniProt | |||||
Find proteins for Q9SNB4 (Arabidopsis thaliana) Explore Q9SNB4 Go to UniProtKB: Q9SNB4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9SNB4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*AP*TP*GP*CP*GP*TP*AP*TP*CP*TP*TP*AP*GP*AP*TP*AP*CP*GP*CP*A)-3') | B [auth U] | 10 | Arabidopsis thaliana | 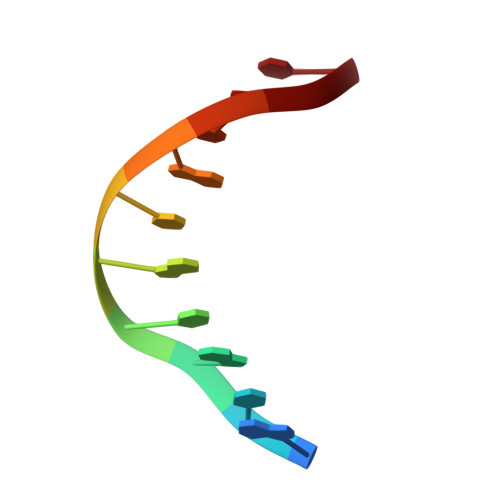 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*AP*TP*GP*CP*GP*TP*AP*TP*CP*TP*TP*AP*GP*AP*TP*AP*CP*GP*CP*A)-3') | C [auth B] | 10 | Arabidopsis thaliana | 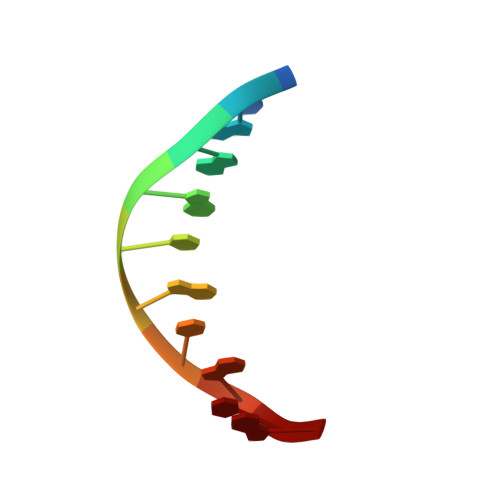 | |
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42.16 | α = 90 |
| b = 32.83 | β = 98.61 |
| c = 53.76 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XSCALE | data scaling |
| SHELX | phasing |
| BUSTER-TNT | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| ATIP-Avenir | France | -- |