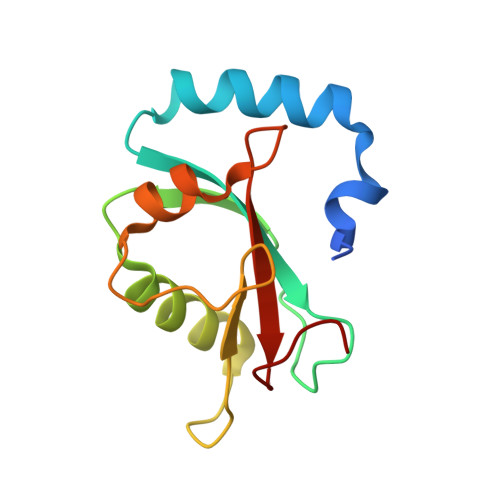
ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Skytte Rasmussen, M., Mouilleron, S., Kumar Shrestha, B., Wirth, M., Lee, R., Bowitz Larsen, K., Abudu Princely, Y., O'Reilly, N., Sjttem, E., Tooze, S.A., Lamark, T., Johansen, T.(2017) Autophagy 13: 834-853
- PubMed: 28287329
- DOI: https://doi.org/10.1080/15548627.2017.1287651
- Primary Citation of Related Structures:
5LXH, 5LXI - PubMed Abstract:
The cysteine protease ATG4B cleaves off one or more C-terminal residues of the inactive proform of proteins of the ortholog and paralog LC3 and GABARAP subfamilies of yeast Atg8 to expose a C-terminal glycine that is conjugated to phosphatidylethanolamine during autophagosome formation. We show that ATG4B contains a C-terminal LC3-interacting region (LIR) motif important for efficient binding to and cleavage of LC3 and GABARAP proteins. We solved the crystal structures of the GABARAPL1-ATG4B C-terminal LIR complex. Analyses of the structures and in vitro binding assays, using specific point mutants, clearly showed that the ATG4B LIR binds via electrostatic-, aromatic HP1 and hydrophobic HP2 pocket interactions. Both these interactions and the catalytic site-substrate interaction contribute to binding between LC3s or GABARAPs and ATG4B. We also reveal an unexpected role for ATG4B in stabilizing the unlipidated forms of GABARAP and GABARAPL1. In mouse embryonic fibroblast (MEF) atg4b knockout cells, GABARAP and GABARAPL1 were unstable and degraded by the proteasome. Strikingly, the LIR motif of ATG4B was required for stabilization of the unlipidated forms of GABARAP and GABARAPL1 in cells.
- a Molecular Cancer Research Group, Department of Medical Biology , University of Tromsø -The Arctic University of Norway , Tromsø , Norway.
Organizational Affiliation: