A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin.
Sgromo, A., Raisch, T., Bawankar, P., Bhandari, D., Chen, Y., Kuzuoglu-Ozturk, D., Weichenrieder, O., Izaurralde, E.(2017) Nat Commun 8: 14307-14307
- PubMed: 28165457
- DOI: https://doi.org/10.1038/ncomms14307
- Primary Citation of Related Structures:
5LSW - PubMed Abstract:
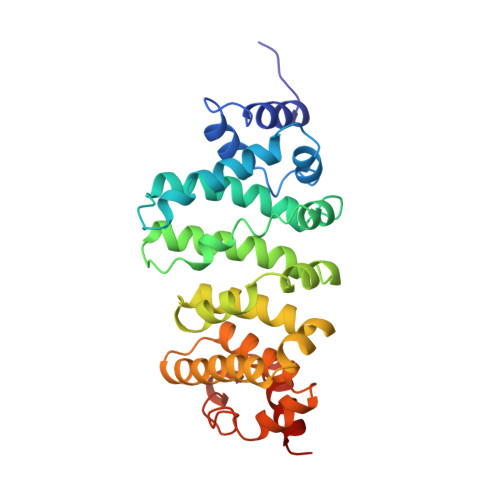
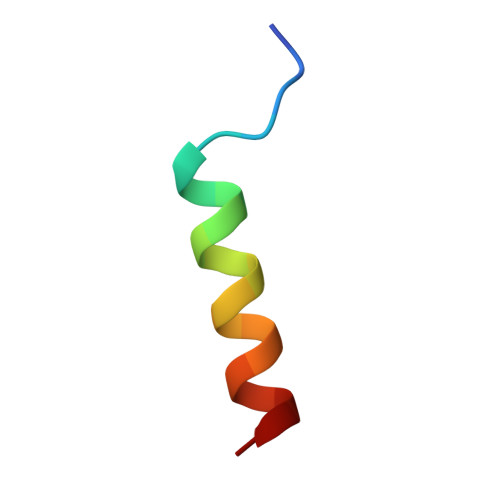
Human (Hs) Roquin1 and Roquin2 are RNA-binding proteins that promote mRNA target degradation through the recruitment of the CCR4-NOT deadenylase complex and are implicated in the prevention of autoimmunity. Roquin1 recruits CCR4-NOT via a C-terminal region that is not conserved in Roquin2 or in invertebrate Roquin. Here we show that Roquin2 and Drosophila melanogaster (Dm) Roquin also interact with the CCR4-NOT complex through their C-terminal regions. The C-terminal region of Dm Roquin contains multiple motifs that mediate CCR4-NOT binding. One motif binds to the CAF40 subunit of the CCR4-NOT complex. The crystal structure of the Dm Roquin CAF40-binding motif (CBM) bound to CAF40 reveals that the CBM adopts an α-helical conformation upon binding to a conserved surface of CAF40. Thus, despite the lack of sequence conservation, the C-terminal regions of Roquin proteins act as an effector domain that represses the expression of mRNA targets via recruitment of the CCR4-NOT complex.
- Department of Biochemistry, Max Planck Institute for Developmental Biology, Spemannstrasse 35, Tübingen 72076, Germany.
Organizational Affiliation: