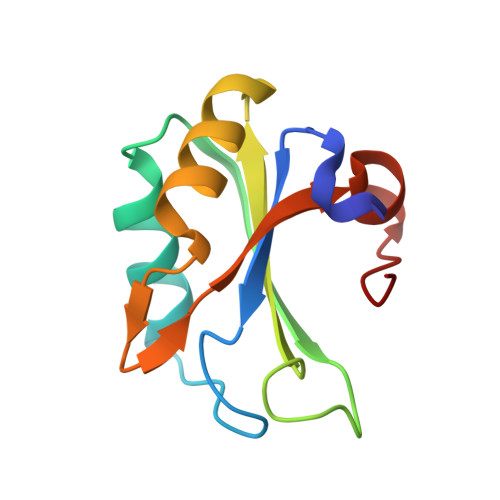
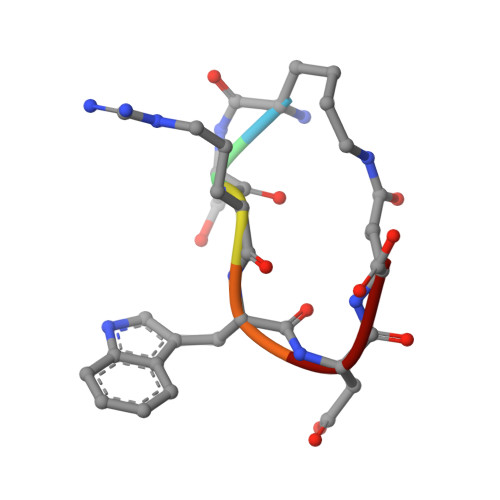
Rational Design of Cyclic Peptide Inhibitors of U2AF Homology Motif (UHM) Domains To Modulate Pre-mRNA Splicing.
Jagtap, P.K., Garg, D., Kapp, T.G., Will, C.L., Demmer, O., Luhrmann, R., Kessler, H., Sattler, M.(2016) J Med Chem 59: 10190-10197
- PubMed: 27753493
- DOI: https://doi.org/10.1021/acs.jmedchem.6b01118
- Primary Citation of Related Structures:
5LSO - PubMed Abstract:
U2AF homology motifs (UHMs) are atypical RNA recognition motif domains that mediate critical protein-protein interactions during the regulation of alternative pre-mRNA splicing and other processes. The recognition of UHM domains by UHM ligand motif (ULM) peptide sequences plays important roles during early steps of spliceosome assembly. Splicing factor 45 kDa (SPF45) is an alternative splicing factor implicated in breast and lung cancers, and splicing regulation of apoptosis-linked pre-mRNAs by SPF45 was shown to depend on interactions between its UHM domain and ULM motifs in constitutive splicing factors. We have developed cyclic peptide inhibitors that target UHM domains. By screening a focused library of linear and cyclic peptides and performing structure-activity relationship analysis, we designed cyclic peptides with 4-fold improved binding affinity for the SPF45 UHM domain compared to native ULM ligands and 270-fold selectivity to discriminate UHM domains from alternative and constitutive splicing factors. These inhibitors are useful tools to modulate and dissect mechanisms of alternative splicing regulation.
- Institute of Structural Biology, Helmholtz Zentrum München , Ingolstaedter Landstrasse 1, 85764 Neuherberg, Germany.
Organizational Affiliation: