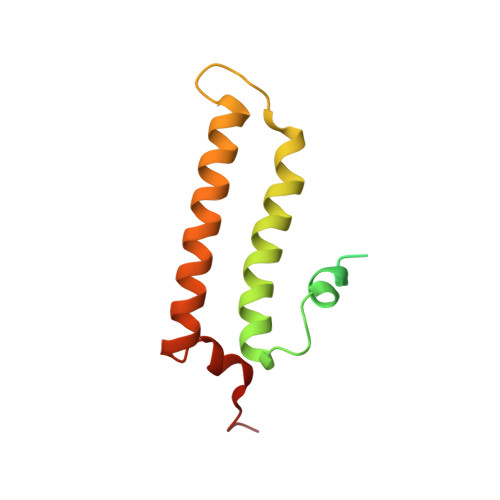
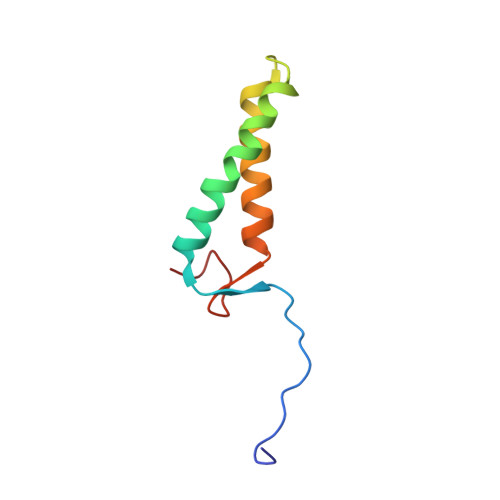
Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Petrovic, A., Keller, J., Liu, Y., Overlack, K., John, J., Dimitrova, Y.N., Jenni, S., van Gerwen, S., Stege, P., Wohlgemuth, S., Rombaut, P., Herzog, F., Harrison, S.C., Vetter, I.R., Musacchio, A.(2016) Cell 167: 1028-1040.e15
- PubMed: 27881301
- DOI: https://doi.org/10.1016/j.cell.2016.10.005
- Primary Citation of Related Structures:
5LSI, 5LSJ, 5LSK - PubMed Abstract:
Kinetochores, multisubunit protein assemblies, connect chromosomes to spindle microtubules to promote chromosome segregation. The 10-subunit KMN assembly (comprising KNL1, MIS12, and NDC80 complexes, designated KNL1C, MIS12C, and NDC80C) binds microtubules and regulates mitotic checkpoint function through NDC80C and KNL1C, respectively. MIS12C, on the other hand, connects the KMN to the chromosome-proximal domain of the kinetochore through a direct interaction with CENP-C. The structural basis for this crucial bridging function of MIS12C is unknown. Here, we report crystal structures of human MIS12C associated with a fragment of CENP-C and unveil the role of Aurora B kinase in the regulation of this interaction. The structure of MIS12:CENP-C complements previously determined high-resolution structures of functional regions of NDC80C and KNL1C and allows us to build a near-complete structural model of the KMN assembly. Our work illuminates the structural organization of essential chromosome segregation machinery that is conserved in most eukaryotes.
- Department of Mechanistic Cell Biology, Max Planck Institute of Molecular Physiology, Otto-Hahn-Straße 11, 44227 Dortmund, Germany. Electronic address: arsen.petrovic@mpi-dortmund.mpg.de.
Organizational Affiliation: