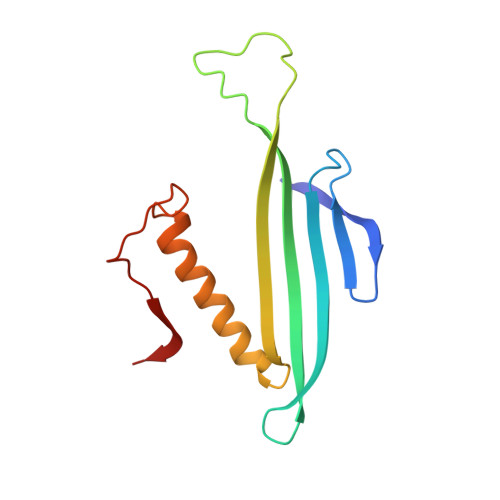
Structure of AP205 Coat Protein Reveals Circular Permutation in ssRNA Bacteriophages.
Shishovs, M., Rumnieks, J., Diebolder, C., Jaudzems, K., Andreas, L.B., Stanek, J., Kazaks, A., Kotelovica, S., Akopjana, I., Pintacuda, G., Koning, R.I., Tars, K.(2016) J Mol Biology 428: 4267-4279
- PubMed: 27591890
- DOI: https://doi.org/10.1016/j.jmb.2016.08.025
- Primary Citation of Related Structures:
5FS4, 5LQP - PubMed Abstract:
AP205 is a single-stranded RNA bacteriophage that has a coat protein sequence not similar to any other known single-stranded RNA phage. Here, we report an atomic-resolution model of the AP205 virus-like particle based on a crystal structure of an unassembled coat protein dimer and a cryo-electron microscopy reconstruction of the assembled particle, together with secondary structure information from site-specific solid-state NMR data. The AP205 coat protein dimer adopts the conserved Leviviridae coat protein fold except for the N-terminal region, which forms a beta-hairpin in the other known single-stranded RNA phages. AP205 has a similar structure at the same location formed by N- and C-terminal beta-strands, making it a circular permutant compared to the other coat proteins. The permutation moves the coat protein termini to the most surface-exposed part of the assembled particle, which explains its increased tolerance to long N- and C-terminal fusions.
- Latvian Biomedical Research and Study Center, Rātsupītes 1, LV1067 Riga, Latvia.
Organizational Affiliation: