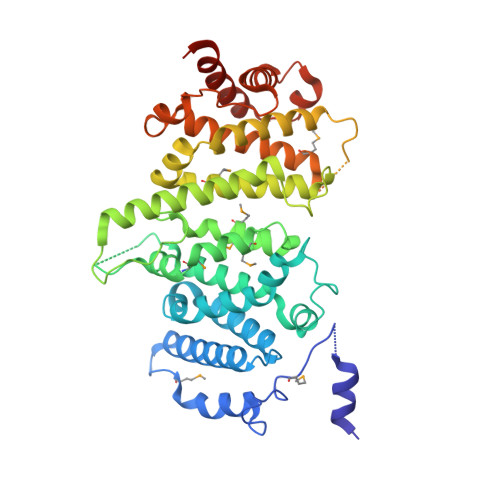
Insights into Rad3 kinase recruitment from the crystal structure of the DNA damage checkpoint protein Rad26.
Andersen, K.R.(2017) J Biological Chem 292: 8149-8157
- PubMed: 28314775
- DOI: https://doi.org/10.1074/jbc.M117.780189
- Primary Citation of Related Structures:
5LOI - PubMed Abstract:
Metabolic products and environmental factors constantly damage DNA. To protect against these insults and maintain genome integrity, cells have evolved mechanisms to repair DNA lesions. One such mechanism involves Rad3, a master kinase coordinating the DNA damage response. Rad26 is a functional subunit of the Rad3-Rad26 complex and is responsible for bringing the kinase to sites of DNA damage. Here, I present the crystal structure of Rad26 and identify the elements important for recruiting Rad3. The structure suggests that Rad26 is a dimer with a conserved interface in the N-terminal part of the protein. Biochemical data showed that Rad26 uses its C-terminal domain and the flanking kinase-docking motif to bind specific HEAT repeats in Rad3. Analysis of the reconstituted Rad3-Rad26 heterotetrameric complex with electron microscopy enabled me to propose a structural model for its quaternary structure. In conclusion, these results suggest that Rad26 exists as a dimer and provide crucial insight into how Rad3 is recruited and incorporated into the Rad3-Rad26 DNA repair complex.
- Department of Molecular Biology and Genetics, Aarhus University, 8000 Aarhus C, Denmark. Electronic address: kra@mbg.au.dk.
Organizational Affiliation: