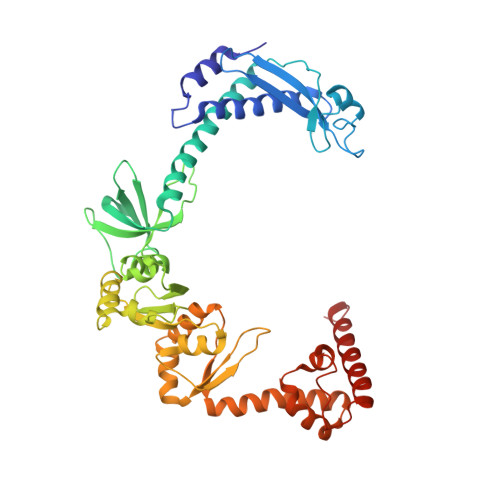
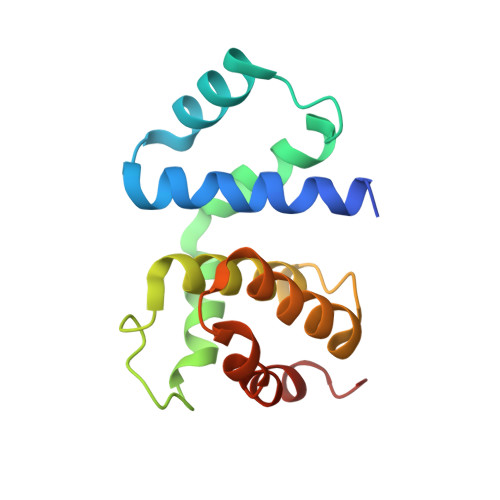
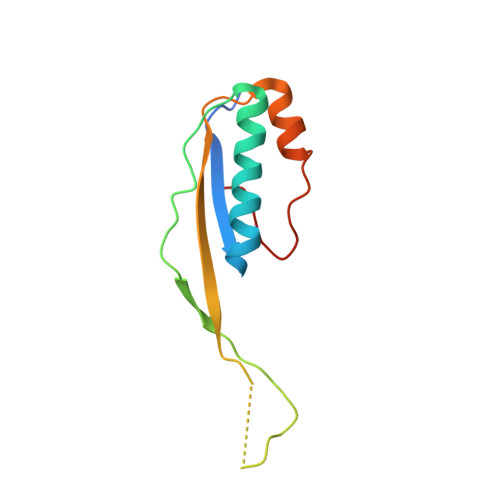
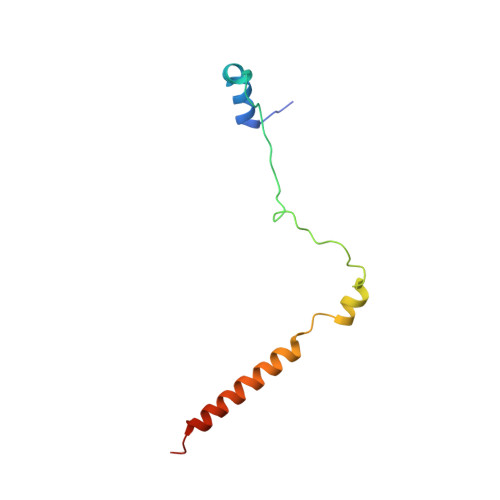
Structural basis for lambda N-dependent processive transcription antitermination.
Said, N., Krupp, F., Anedchenko, E., Santos, K.F., Dybkov, O., Huang, Y.H., Lee, C.T., Loll, B., Behrmann, E., Burger, J., Mielke, T., Loerke, J., Urlaub, H., Spahn, C.M.T., Weber, G., Wahl, M.C.(2017) Nat Microbiol 2: 17062-17062
- PubMed: 28452979
- DOI: https://doi.org/10.1038/nmicrobiol.2017.62
- Primary Citation of Related Structures:
5LM7, 5LM9, 5MS0 - PubMed Abstract:
λN-mediated processive antitermination constitutes a paradigmatic transcription regulatory event, during which phage protein λN, host factors NusA, NusB, NusE and NusG, and an RNA nut site render elongating RNA polymerase termination-resistant. The structural basis of the process has so far remained elusive. Here we describe a crystal structure of a λN-NusA-NusB-NusE-nut site complex and an electron cryo-microscopic structure of a complete transcription antitermination complex, comprising RNA polymerase, DNA, nut site RNA, all Nus factors and λN, validated by crosslinking/mass spectrometry. Due to intrinsic disorder, λN can act as a multiprotein/RNA interaction hub, which, together with nut site RNA, arranges NusA, NusB and NusE into a triangular complex. This complex docks via the NusA N-terminal domain and the λN C-terminus next to the RNA exit channel on RNA polymerase. Based on the structures, comparative crosslinking analyses and structure-guided mutagenesis, we hypothesize that λN mounts a multipronged strategy to reprogram the transcriptional machinery, which may include (1) the λN C terminus clamping the RNA exit channel, thus stabilizing the DNA:RNA hybrid; (2) repositioning of NusA and RNAP elements, thus redirecting nascent RNA and sequestering the upstream branch of a terminator hairpin; and (3) hindering RNA engagement of termination factor ρ and/or obstructing ρ translocation on the transcript.
- Laboratory of Structural Biochemistry, Freie Universität Berlin, Takustraβe 6, D-14195 Berlin, Germany.
Organizational Affiliation: