Mechanistic Insight into Bunyavirus-Induced Membrane Fusion from Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc.
Guardado-Calvo, P., Bignon, E.A., Stettner, E., Jeffers, S.A., Perez-Vargas, J., Pehau-Arnaudet, G., Tortorici, M.A., Jestin, J.L., England, P., Tischler, N.D., Rey, F.A.(2016) PLoS Pathog 12: e1005813-e1005813
- PubMed: 27783711
- DOI: https://doi.org/10.1371/journal.ppat.1005813
- Primary Citation of Related Structures:
5LJX, 5LJY, 5LJZ, 5LK0, 5LK1, 5LK2, 5LK3 - PubMed Abstract:
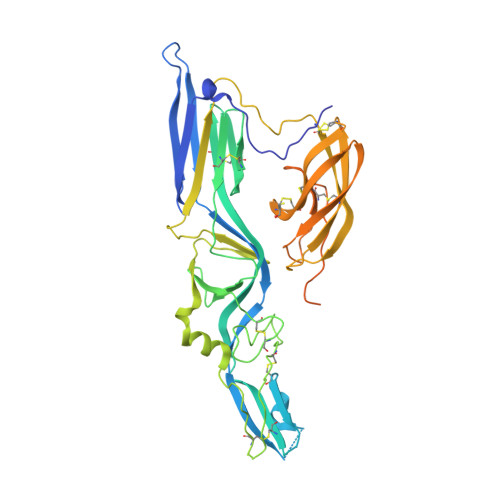
Hantaviruses are zoonotic viruses transmitted to humans by persistently infected rodents, giving rise to serious outbreaks of hemorrhagic fever with renal syndrome (HFRS) or of hantavirus pulmonary syndrome (HPS), depending on the virus, which are associated with high case fatality rates. There is only limited knowledge about the organization of the viral particles and in particular, about the hantavirus membrane fusion glycoprotein Gc, the function of which is essential for virus entry. We describe here the X-ray structures of Gc from Hantaan virus, the type species hantavirus and responsible for HFRS, both in its neutral pH, monomeric pre-fusion conformation, and in its acidic pH, trimeric post-fusion form. The structures confirm the prediction that Gc is a class II fusion protein, containing the characteristic β-sheet rich domains termed I, II and III as initially identified in the fusion proteins of arboviruses such as alpha- and flaviviruses. The structures also show a number of features of Gc that are distinct from arbovirus class II proteins. In particular, hantavirus Gc inserts residues from three different loops into the target membrane to drive fusion, as confirmed functionally by structure-guided mutagenesis on the HPS-inducing Andes virus, instead of having a single "fusion loop". We further show that the membrane interacting region of Gc becomes structured only at acidic pH via a set of polar and electrostatic interactions. Furthermore, the structure reveals that hantavirus Gc has an additional N-terminal "tail" that is crucial in stabilizing the post-fusion trimer, accompanying the swapping of domain III in the quaternary arrangement of the trimer as compared to the standard class II fusion proteins. The mechanistic understandings derived from these data are likely to provide a unique handle for devising treatments against these human pathogens.
- Institut Pasteur, Unité de Virologie Structurale, Département de Virologie, Paris, France.
Organizational Affiliation: