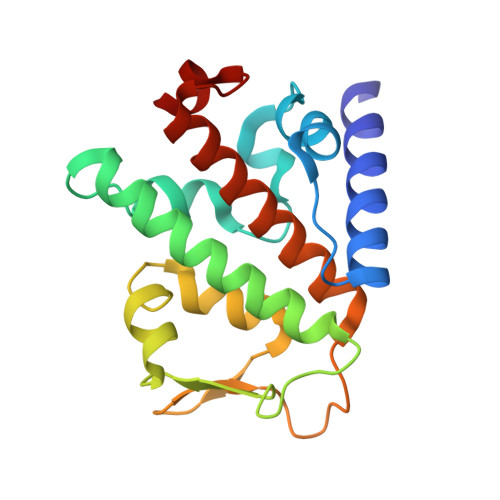
SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Elliott, P.R., Leske, D., Hrdinka, M., Bagola, K., Fiil, B.K., McLaughlin, S.H., Wagstaff, J., Volkmar, N., Christianson, J.C., Kessler, B.M., Freund, S.M., Komander, D., Gyrd-Hansen, M.(2016) Mol Cell 63: 990-1005
- PubMed: 27591049
- DOI: https://doi.org/10.1016/j.molcel.2016.08.001
- Primary Citation of Related Structures:
5LJM, 5LJN - PubMed Abstract:
The linear ubiquitin chain assembly complex (LUBAC) regulates immune signaling, and its function is regulated by the deubiquitinases OTULIN and CYLD, which associate with the catalytic subunit HOIP. However, the mechanism through which CYLD interacts with HOIP is unclear. We here show that CYLD interacts with HOIP via spermatogenesis-associated protein 2 (SPATA2). SPATA2 interacts with CYLD through its non-canonical PUB domain, which binds the catalytic CYLD USP domain in a CYLD B-box-dependent manner. Significantly, SPATA2 binding activates CYLD-mediated hydrolysis of ubiquitin chains. SPATA2 also harbors a conserved PUB-interacting motif that selectively docks into the HOIP PUB domain. In cells, SPATA2 is recruited to the TNF receptor 1 signaling complex and is required for CYLD recruitment. Loss of SPATA2 increases ubiquitination of LUBAC substrates and results in enhanced NOD2 signaling. Our data reveal SPATA2 as a high-affinity binding partner of CYLD and HOIP, and a regulatory component of LUBAC-mediated NF-κB signaling.
- Division of Protein and Nucleic Acid Chemistry, MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge CB2 0QH, UK.
Organizational Affiliation: