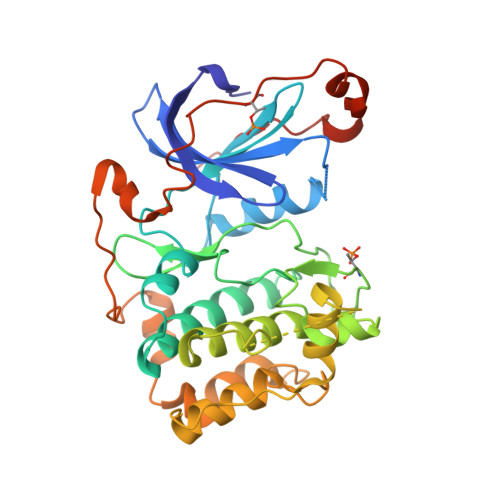
aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Soriano, E.V., Ivanova, M.E., Fletcher, G., Riou, P., Knowles, P.P., Barnouin, K., Purkiss, A., Kostelecky, B., Saiu, P., Linch, M., Elbediwy, A., Kjr, S., O'Reilly, N., Snijders, A.P., Parker, P.J., Thompson, B.J., McDonald, N.Q.(2016) Dev Cell 38: 384-398
- PubMed: 27554858
- DOI: https://doi.org/10.1016/j.devcel.2016.07.018
- Primary Citation of Related Structures:
5LI1, 5LI9, 5LIH - PubMed Abstract:
Atypical protein kinase C (aPKC) is a key apical-basal polarity determinant and Par complex component. It is recruited by Par3/Baz (Bazooka in Drosophila) into epithelial apical domains through high-affinity interaction. Paradoxically, aPKC also phosphorylates Par3/Baz, provoking its relocalization to adherens junctions (AJs). We show that Par3 conserved region 3 (CR3) forms a tight inhibitory complex with a primed aPKC kinase domain, blocking substrate access. A CR3 motif flanking its PKC consensus site disrupts the aPKC kinase N lobe, separating P-loop/αB/αC contacts. A second CR3 motif provides a high-affinity anchor. Mutation of either motif switches CR3 to an efficient in vitro substrate by exposing its phospho-acceptor site. In vivo, mutation of either CR3 motif alters Par3/Baz localization from apical to AJs. Our results reveal how Par3/Baz CR3 can antagonize aPKC in stable apical Par complexes and suggests that modulation of CR3 inhibitory arms or opposing aPKC pockets would perturb the interaction, promoting Par3/Baz phosphorylation.
- Structural Biology, The Francis Crick Institute, 44 Lincoln's Inn Fields, London WC2A 3LY, UK.
Organizational Affiliation: