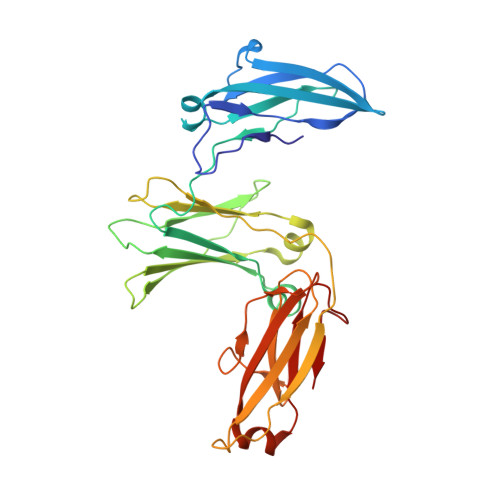
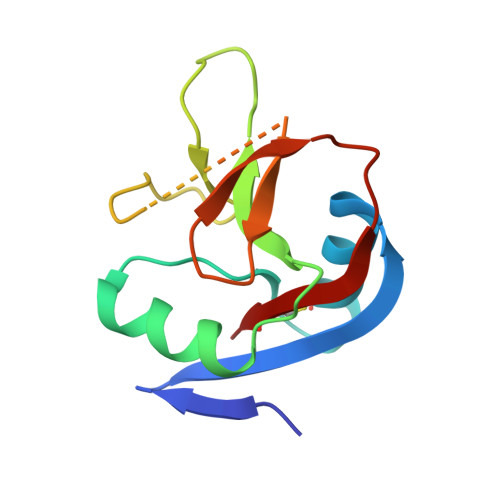
IgE binds asymmetrically to its B cell receptor CD23.
Dhaliwal, B., Pang, M.O., Keeble, A.H., James, L.K., Gould, H.J., McDonnell, J.M., Sutton, B.J., Beavil, A.J.(2017) Sci Rep 7: 45533-45533
- PubMed: 28361904
- DOI: https://doi.org/10.1038/srep45533
- Primary Citation of Related Structures:
5LGK - PubMed Abstract:
The antibody IgE plays a central role in allergic disease mechanisms. Its effector functions are controlled through interactions between the Fc region and two principal cell surface receptors FcεRI and CD23. The interaction with FcεRI is primarily responsible for allergic sensitization and the inflammatory response, while IgE binding to CD23 is involved in the regulation of IgE synthesis and allergen transcytosis. Here we present the crystal structure of a CD23/IgE-Fc complex and conduct isothermal titration calorimetric binding studies. Two lectin-like "head" domains of CD23 bind to IgE-Fc with affinities that differ by more than an order of magnitude, but the crystal structure reveals only one head bound to one of the two identical heavy-chains in the asymmetrically bent IgE-Fc. These results highlight the subtle interplay between receptor binding sites in IgE-Fc and their affinities, the understanding of which may be exploited for therapeutic intervention in allergic disease.
- Division of Infection, Immunity and Respiratory Medicine, School of Biological Sciences, Faculty of Biology, Medicine and Health, Manchester Academic Health Sciences Centre and Manchester Institute of Biotechnology, University of Manchester, 131 Princess Street, Manchester, M1 7DN, U.K.
Organizational Affiliation: