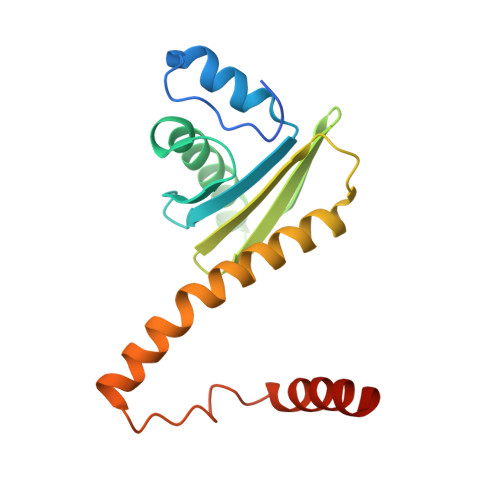
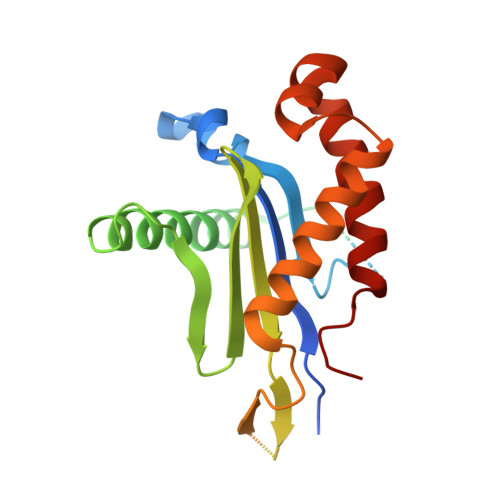
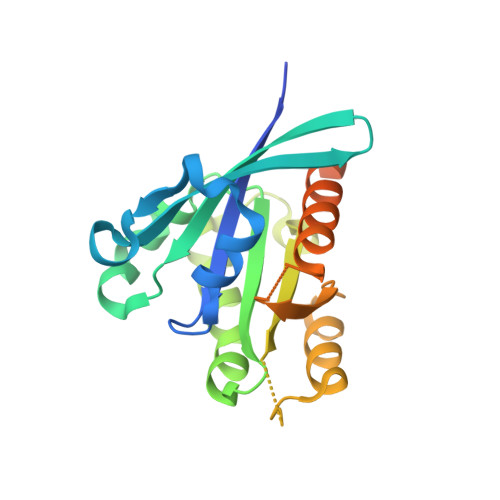
Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1.
Kiontke, S., Langemeyer, L., Kuhlee, A., Schuback, S., Raunser, S., Ungermann, C., Kummel, D.(2017) Nat Commun 8: 14034-14034
- PubMed: 28051187
- DOI: https://doi.org/10.1038/ncomms14034
- Primary Citation of Related Structures:
5LDD - PubMed Abstract:
The Mon1-Ccz1 complex (MC1) is the guanine nucleotide exchange factor (GEF) for the Rab GTPase Ypt7/Rab7 and is required for endosomal maturation and fusion at the vacuole/lysosome. Here we present the overall architecture of MC1 from Chaetomium thermophilum, and in combining biochemical studies and mutational analysis in yeast, we identify the domains required for catalytic activity, complex assembly and localization of MC1. The crystal structure of a catalytic MC1 core complex bound to Ypt7 provides mechanistic insight into its function. We pinpoint the determinants that allow for a discrimination of the Rab7-like Ypt7 over the Rab5-like Vps21, which are both located on the same membrane. MC1 shares structural similarities with the TRAPP complex, but employs a novel mechanism to promote nucleotide exchange that utilizes a conserved lysine residue of Ypt7, which is inserted upon MC1 binding into the nucleotide-binding pocket of Ypt7 and contributes to specificity.
- Division of Structural Biology, Department of Biology/Chemistry, University of Osnabrück, Barbarastraße 13, Osnabrück 49076, Germany.
Organizational Affiliation: