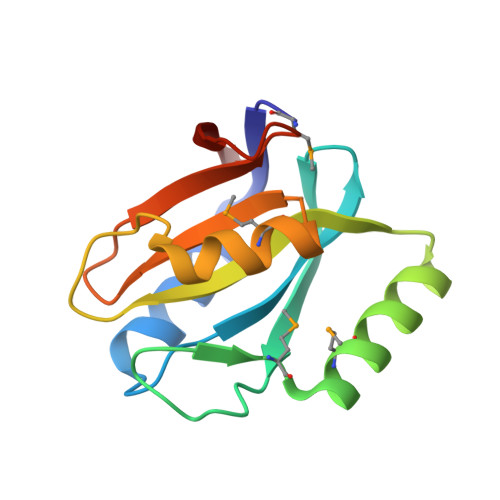
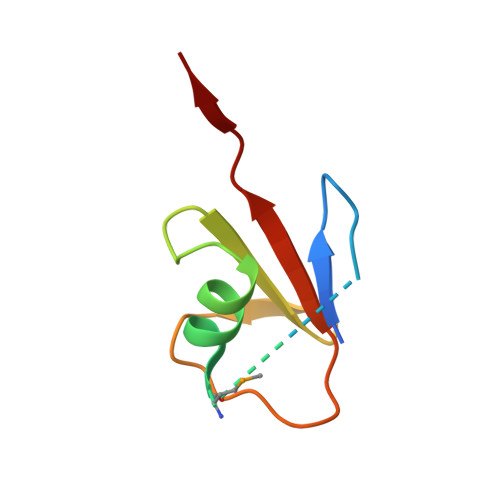
Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Cao, S., Engilberge, S., Girard, E., Gabel, F., Franzetti, B., Maupin-Furlow, J.A.(2017) Structure 25: 823-833.e6
- PubMed: 28479062
- DOI: https://doi.org/10.1016/j.str.2017.04.002
- Primary Citation of Related Structures:
5LD9, 5LDA - PubMed Abstract:
JAMM/MPN + metalloproteases cleave (iso)peptide bonds C-terminal to ubiquitin (Ub) and ubiquitin-like protein (Ubl) domains and typically require association with protein partners for activity, which has limited a molecular understanding of enzyme function. To provide an insight, we solved the X-ray crystal structures of a catalytically active Pyrococcus furiosus JAMM/MPN + metalloprotease (PfJAMM1) alone and in complex with a Ubl (PfSAMP2) to 1.7- to 1.9-Å resolution. PfJAMM1 was found to have a redox sensitive dimer interface. In the PfJAMM1-bound state of the SAMP2, a Ubl-to-Ub conformational change was detected. Surprisingly, distant homologs of PfJAMM1 were found to be closely related in 3D structure, including the interface for Ubl/Ub binding. From this work, we infer the molecular basis of how JAMM/MPN + proteases recognize and cleave Ubl/Ub tags from diverse proteins and highlight an α2-helix structural element that is conserved and crucial for binding and removing the Ubl SAMP2 tag.
- Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL 32611, USA.
Organizational Affiliation: