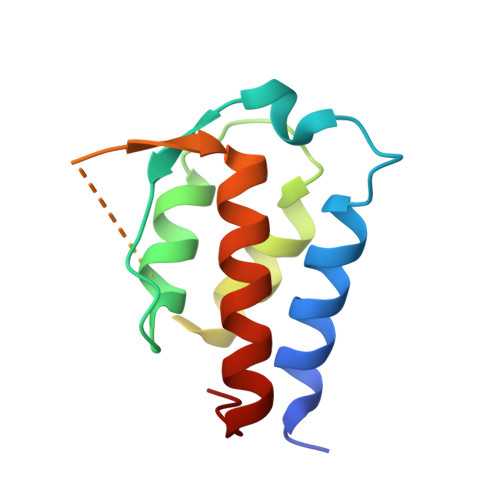
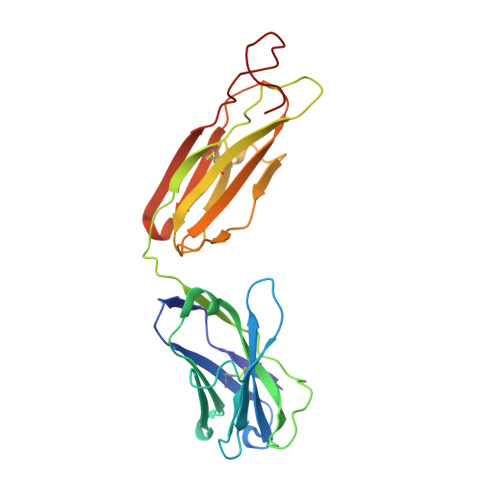
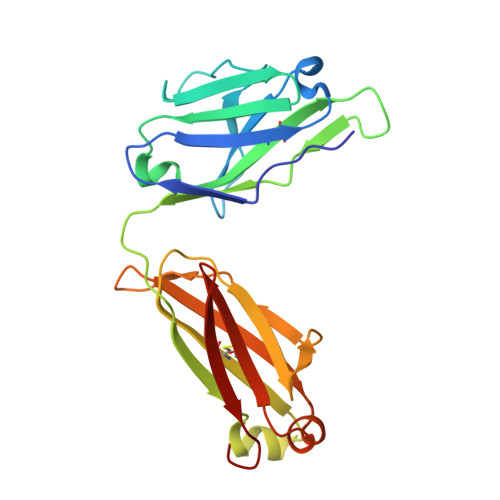
Structural Characterisation Reveals Mechanism of IL-13-Neutralising Monoclonal Antibody Tralokinumab as Inhibition of Binding to IL-13R alpha 1 and IL-13R alpha 2.
Popovic, B., Breed, J., Rees, D.G., Gardener, M.J., Vinall, L.M., Kemp, B., Spooner, J., Keen, J., Minter, R., Uddin, F., Colice, G., Wilkinson, T., Vaughan, T., May, R.D.(2017) J Mol Biology 429: 208-219
- PubMed: 27956146
- DOI: https://doi.org/10.1016/j.jmb.2016.12.005
- Primary Citation of Related Structures:
5L6Y - PubMed Abstract:
Interleukin (IL)-13 is a pleiotropic T helper type 2 cytokine frequently associated with asthma and atopic dermatitis. IL-13-mediated signalling is initiated by binding to IL-13Rα1, which then recruits IL-4Rα to form a heterodimeric receptor complex. IL-13 also binds to IL-13Rα2, considered as either a decoy or a key mediator of fibrosis. IL-13-neutralising antibodies act by preventing IL-13 binding to IL-13Rα1, IL-4Rα and/or IL-13Rα2. Tralokinumab (CAT-354) is an IL-13-neutralising human IgG4 monoclonal antibody that has shown clinical benefit in patients with asthma. To decipher how tralokinumab inhibits the effects of IL-13, we determined the structure of tralokinumab Fab in complex with human IL-13 to 2 Å resolution. The structure analysis reveals that tralokinumab prevents IL-13 from binding to both IL-13Rα1 and IL-13Rα2. This is supported by biochemical ligand-receptor interaction assay data. The tralokinumab epitope is mainly composed of residues in helices D and A of IL-13. It is mostly light chain complementarity-determining regions that are driving paratope interactions; the variable light complementarity-determining region 2 plays a key role by providing residue contacts for a network of hydrogen bonds and a salt bridge in the core of binding. The key residues within the paratope contributing to binding were identified as Asp50, Asp51, Ser30 and Lys31. This study demonstrates that tralokinumab prevents the IL-13 pharmacodynamic effect by binding to IL-13 helices A and D, thus preventing IL-13 from interacting with IL-13Rα1 and IL-13Rα2.
- Department of Antibody Discovery and Protein Engineering, MedImmune Ltd., Granta Park, Cambridge CB21 6GH, UK. Electronic address: popovicb@medimmune.com.
Organizational Affiliation: