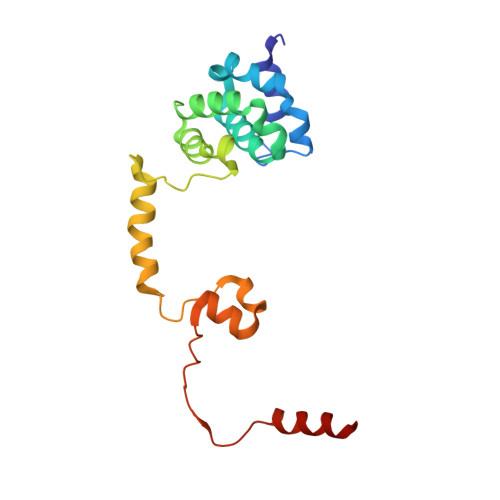
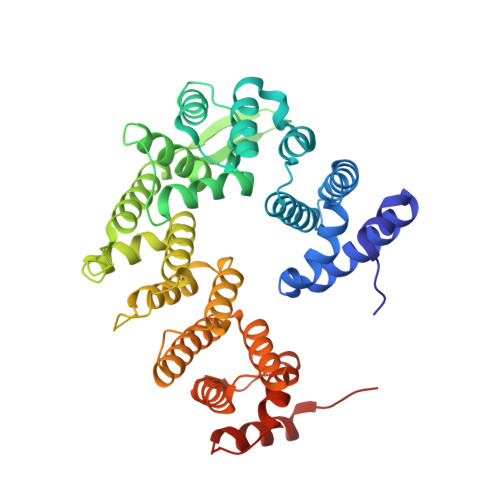
Architecture and RNA binding of the human negative elongation factor.
Vos, S.M., Pollmann, D., Caizzi, L., Hofmann, K.B., Rombaut, P., Zimniak, T., Herzog, F., Cramer, P.(2016) Elife 5
- PubMed: 27282391
- DOI: https://doi.org/10.7554/eLife.14981
- Primary Citation of Related Structures:
5L3X - PubMed Abstract:
Transcription regulation in metazoans often involves promoter-proximal pausing of RNA polymerase (Pol) II, which requires the 4-subunit negative elongation factor (NELF). Here we discern the functional architecture of human NELF through X-ray crystallography, protein crosslinking, biochemical assays, and RNA crosslinking in cells. We identify a NELF core subcomplex formed by conserved regions in subunits NELF-A and NELF-C, and resolve its crystal structure. The NELF-AC subcomplex binds single-stranded nucleic acids in vitro, and NELF-C associates with RNA in vivo. A positively charged face of NELF-AC is involved in RNA binding, whereas the opposite face of the NELF-AC subcomplex binds NELF-B. NELF-B is predicted to form a HEAT repeat fold, also binds RNA in vivo, and anchors the subunit NELF-E, which is confirmed to bind RNA in vivo. These results reveal the three-dimensional architecture and three RNA-binding faces of NELF.
- Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany.
Organizational Affiliation: