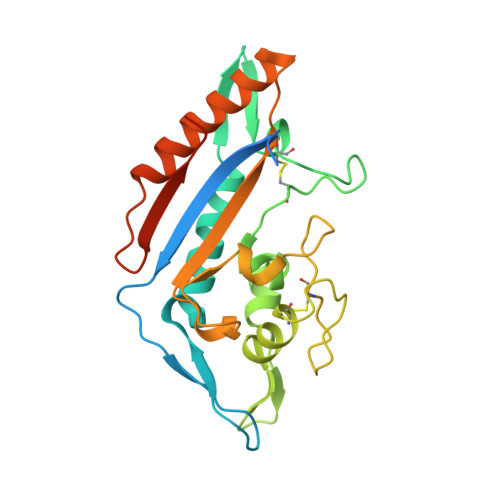
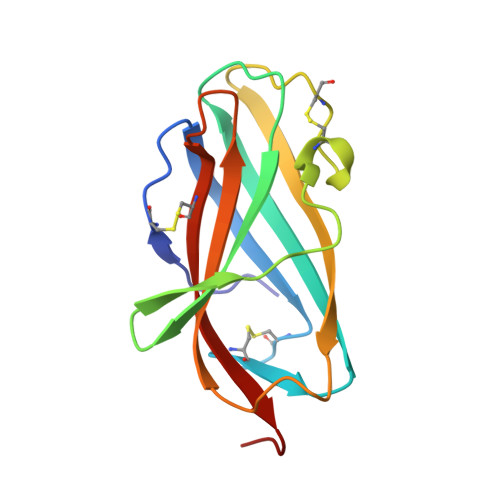
Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2.
Li, X., Saha, P., Li, J., Blobel, G., Pfeffer, S.R.(2016) Proc Natl Acad Sci U S A 113: 10079-10084
- PubMed: 27551080
- DOI: https://doi.org/10.1073/pnas.1611956113
- Primary Citation of Related Structures:
5KWY - PubMed Abstract:
Export of LDL-derived cholesterol from lysosomes requires the cooperation of the integral membrane protein Niemann-Pick C1 (NPC1) and a soluble protein, Niemann-Pick C2 (NPC2). Mutations in the genes encoding these proteins lead to Niemann-Pick disease type C (NPC). NPC2 binds to NPC1's second (middle), lumenally oriented domain (MLD) and transfers cholesterol to NPC1's N-terminal domain (NTD). Here, we report the 2.4-Å resolution crystal structure of a complex of human NPC1-MLD and NPC2 bearing bound cholesterol-3-O-sulfate. NPC1-MLD uses two protruding loops to bind NPC2, analogous to its interaction with the primed Ebola virus glycoprotein. Docking of the NPC1-NPC2 complex onto the full-length NPC1 structure reveals a direct cholesterol transfer tunnel between NPC2 and NTD cholesterol binding pockets, supporting the "hydrophobic hand-off" cholesterol transfer model.
- Laboratory of Cell Biology, Howard Hughes Medical Institute, The Rockefeller University, New York, NY 10065; xli05@rockefeller.edu blobel@rockefeller.edu pfeffer@stanford.edu.
Organizational Affiliation: