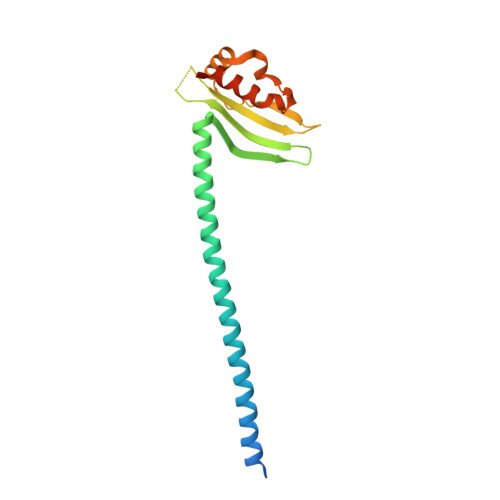
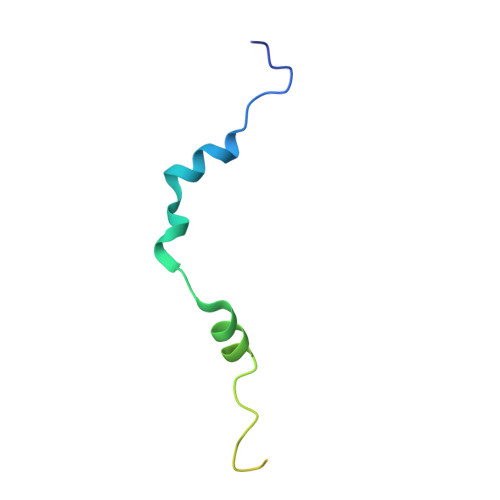
Molecular architecture of the yeast monopolin complex.
Corbett, K.D., Harrison, S.C.(2012) Cell Rep 1: 583-589
- PubMed: 22813733
- DOI: https://doi.org/10.1016/j.celrep.2012.05.012
- Primary Citation of Related Structures:
5KTB - PubMed Abstract:
The Saccharomyces cerevisiae monopolin complex directs proper chromosome segregation in meiosis I by mediating co-orientation of sister kinetochores on the meiosis I spindle. The monopolin subunits Csm1 and Lrs4 form a V-shaped complex that may directly crosslink sister kinetochores. We report here biochemical characterization of the monopolin complex subunits Mam1 and Hrr25 and of the complete four-protein monopolin complex. By purifying monopolin subcomplexes with different subunit combinations, we have determined the stoichiometry and overall architecture of the full monopolin complex. We have determined the crystal structure of Csm1 bound to a Mam1 fragment, showing how Mam1 wraps around the Csm1 dimer and alters the stoichiometry of kinetochore-protein binding by Csm1. We further show that the kinase activity of Hrr25 is altered by Mam1 binding, and we identify Hrr25 phosphorylation sites on Mam1 that may affect monopolin complex stability and/or kinetochore binding in meiosis.
- Ludwig Institute for Cancer Research, Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, CA 92093-0660, USA. kcorbett@ucsd.edu
Organizational Affiliation: