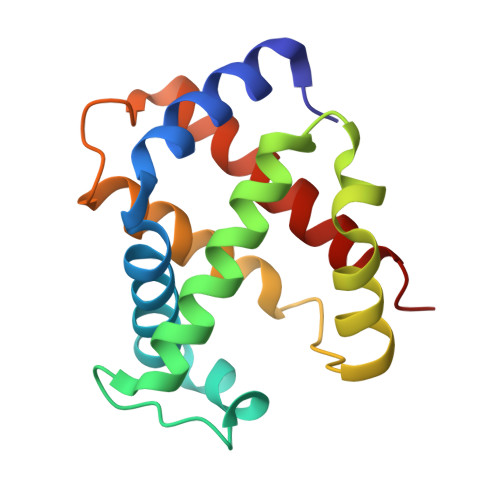
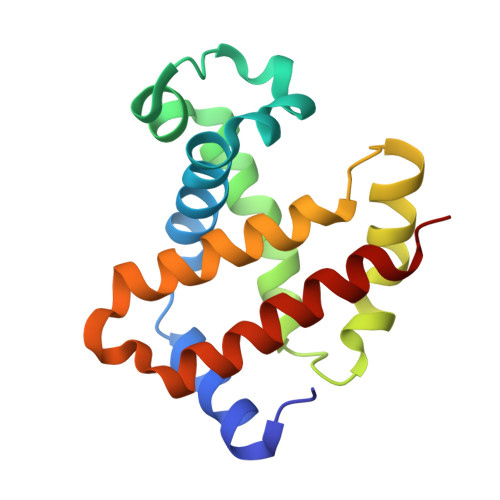
Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sun, K., D'Alessandro, A., Ahmed, M.H., Zhang, Y., Song, A., Ko, T.P., Nemkov, T., Reisz, J.A., Wu, H., Adebiyi, M., Peng, Z., Gong, J., Liu, H., Huang, A., Wen, Y.E., Wen, A.Q., Berka, V., Bogdanov, M.V., Abdulmalik, O., Han, L., Tsai, A.L., Idowu, M., Juneja, H.S., Kellems, R.E., Dowhan, W., Hansen, K.C., Safo, M.K., Xia, Y.(2017) Sci Rep 7: 15281-15281
- PubMed: 29127281
- DOI: https://doi.org/10.1038/s41598-017-13667-8
- Primary Citation of Related Structures:
5KSI, 5KSJ - PubMed Abstract:
Elevated sphingosine 1-phosphate (S1P) is detrimental in Sickle Cell Disease (SCD), but the mechanistic basis remains obscure. Here, we report that increased erythrocyte S1P binds to deoxygenated sickle Hb (deoxyHbS), facilitates deoxyHbS anchoring to the membrane, induces release of membrane-bound glycolytic enzymes and in turn switches glucose flux towards glycolysis relative to the pentose phosphate pathway (PPP). Suppressed PPP causes compromised glutathione homeostasis and increased oxidative stress, while enhanced glycolysis induces production of 2,3-bisphosphoglycerate (2,3-BPG) and thus increases deoxyHbS polymerization, sickling, hemolysis and disease progression. Functional studies revealed that S1P and 2,3-BPG work synergistically to decrease both HbA and HbS oxygen binding affinity. The crystal structure at 1.9 Å resolution deciphered that S1P binds to the surface of 2,3-BPG-deoxyHbA and causes additional conformation changes to the T-state Hb. Phosphate moiety of the surface bound S1P engages in a highly positive region close to α1-heme while its aliphatic chain snakes along a shallow cavity making hydrophobic interactions in the "switch region", as well as with α2-heme like a molecular "sticky tape" with the last 3-4 carbon atoms sticking out into bulk solvent. Altogether, our findings provide functional and structural bases underlying S1P-mediated pathogenic metabolic reprogramming in SCD and novel therapeutic avenues.
- Department of Biochemistry and Molecular Biology, The University of Texas Health Science Center at Houston, Houston, TX, 77030, USA.
Organizational Affiliation: