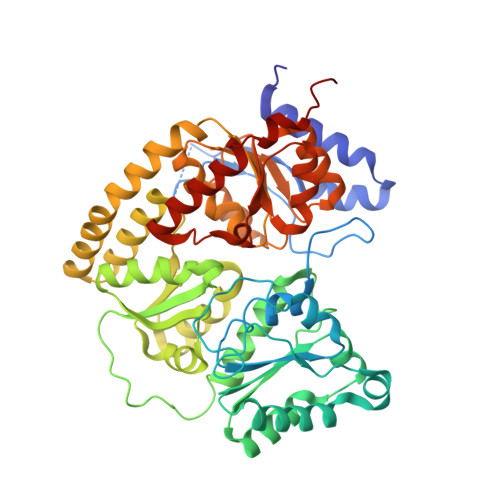
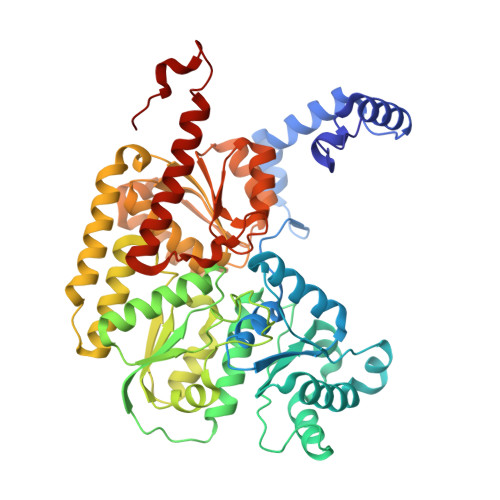
Tyrosine-Coordinated P-Cluster in G. diazotrophicus Nitrogenase: Evidence for the Importance of O-Based Ligands in Conformationally Gated Electron Transfer.
Owens, C.P., Katz, F.E., Carter, C.H., Oswald, V.F., Tezcan, F.A.(2016) J Am Chem Soc 138: 10124-10127
- PubMed: 27487256
- DOI: https://doi.org/10.1021/jacs.6b06783
- Primary Citation of Related Structures:
5KOH, 5KOJ - PubMed Abstract:
The P-cluster is a unique iron-sulfur center that likely functions as a dynamic electron (e(-)) relay site between the Fe-protein and the catalytic FeMo-cofactor in nitrogenase. The P-cluster has been shown to undergo large conformational changes upon 2-e(-) oxidation which entail the coordination of two of the Fe centers to a Ser side chain and a backbone amide N, respectively. Yet, how and if this 2-e(-) oxidized state (P(OX)) is involved in catalysis by nitrogenase is not well established. Here, we present the crystal structures of reduced and oxidized MoFe-protein (MoFeP) from Gluconacetobacter diazotrophicus (Gd), which natively possesses an Ala residue in the position of the Ser ligand to the P-cluster. While reduced Gd-MoFeP is structurally identical to previously characterized counterparts around the FeMo-cofactor, oxidized Gd-MoFeP features an unusual Tyr coordination to its P-cluster along with ligation by a backbone amide nitrogen. EPR analysis of the oxidized Gd-MoFeP P-cluster confirmed that it is a 2-e(-) oxidized, integer-spin species. Importantly, we have found that the sequence positions corresponding to the Ser and Tyr ligands are almost completely covariant among Group I nitrogenases. These findings strongly support the possibility that the P(OX) state is functionally relevant in nitrogenase catalysis and that a hard, O-based anionic ligand serves to stabilize this state in a switchable fashion.
- Department of Chemistry and Biochemistry, University of California, San Diego , 9500 Gilman Drive, La Jolla, California 92093-0356, United States.
Organizational Affiliation: