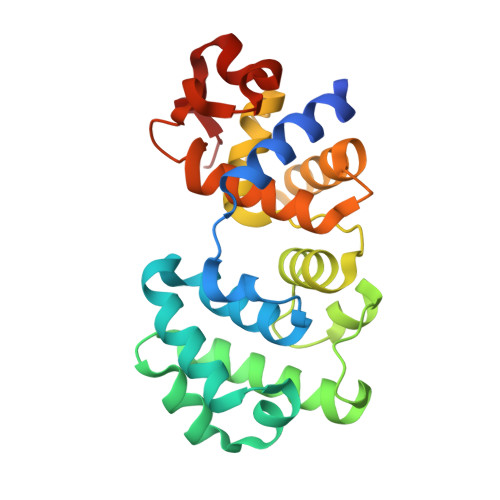
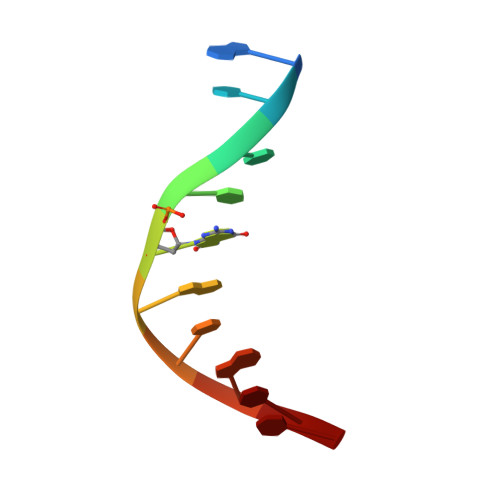
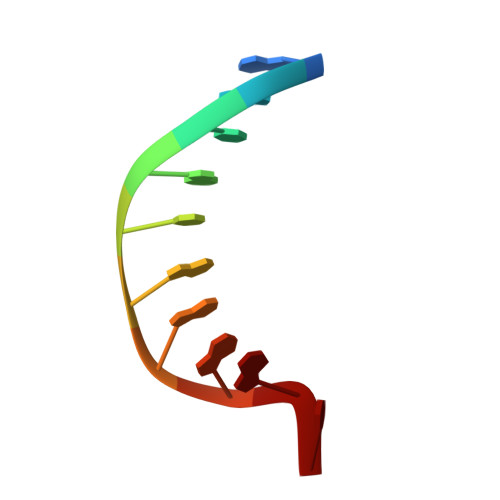
Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
Wang, L., Chakravarthy, S., Verdine, G.L.(2017) J Biological Chem 292: 5007-5017
- PubMed: 28130451
- DOI: https://doi.org/10.1074/jbc.M116.757039
- Primary Citation of Related Structures:
5KN8, 5KN9 - PubMed Abstract:
The highly mutagenic A:8-oxoguanine (oxoG) base pair is generated mainly by misreplication of the C:oxoG base pair, the oxidation product of the C:G base pair. The A:oxoG base pair is particularly insidious because neither base in it carries faithful information to direct the repair of the other. The bacterial MutY (MUTYH in humans) adenine DNA glycosylase is able to initiate the repair of A:oxoG by selectively cleaving the A base from the A:oxoG base pair. The difference between faithful repair and wreaking mutagenic havoc on the genome lies in the accurate discrimination between two structurally similar base pairs: A:oxoG and A:T. Here we present two crystal structures of the MutY N-terminal domain in complex with either undamaged DNA or DNA containing an intrahelical lesion. These structures have captured for the first time a DNA glycosylase scanning the genome for a damaged base in the very first stage of lesion recognition and the base extrusion pathway. The mode of interaction observed here has suggested a common lesion-scanning mechanism across the entire helix-hairpin-helix superfamily to which MutY belongs. In addition, small angle X-ray scattering studies together with accompanying biochemical assays have suggested a possible role played by the C-terminal oxoG-recognition domain of MutY in lesion scanning.
- From the Departments of Chemistry and Chemical Biology.
Organizational Affiliation: