Crystal Structure of SMT Fusion Peptidyl-Prolyl Cis-Trans Isomerase from Burkholderia Pseudomallei Complexed with SF110
Lorimer, D.D., Fox III, D., Seufert, F., Edwards, T.E., Holzgrabe, U.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chimera protein of Ubiquitin-like protein SMT3 and Peptidyl-prolyl cis-trans isomerase | 209 | Saccharomyces cerevisiae S288C, Burkholderia pseudomallei 1710b This entity is chimeric | Mutation(s): 0 Gene Names: SMT3, YDR510W, D9719.15, BURPS1710b_A0907 EC: 5.2.1.8 | 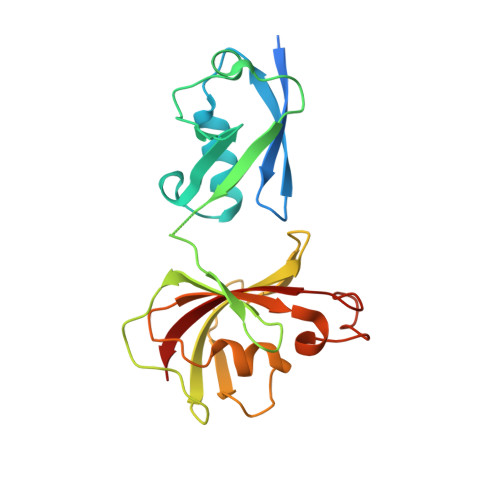 | |
UniProt | |||||
Find proteins for Q12306 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q12306 Go to UniProtKB: Q12306 | |||||
Find proteins for Q3JK38 (Burkholderia pseudomallei (strain 1710b)) Explore Q3JK38 Go to UniProtKB: Q3JK38 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q3JK38Q12306 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 6UO Query on 6UO | E [auth A], I [auth B], J [auth C], K [auth D] | 2-[(2~{S})-1-(phenylmethyl)sulfonylpiperidin-2-yl]carbonyloxyethyl pyridine-3-carboxylate C21 H24 N2 O6 S KUWLHSSIHRTCQU-IBGZPJMESA-N |  | ||
| PEG Query on PEG | H [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| IMD Query on IMD | G [auth A], M [auth D] | IMIDAZOLE C3 H5 N2 RAXXELZNTBOGNW-UHFFFAOYSA-O |  | ||
| MG Query on MG | F [auth A], L [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 107.89 | α = 90 |
| b = 34.11 | β = 115.45 |
| c = 119.92 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| PHENIX | refinement |
| PHENIX | phasing |