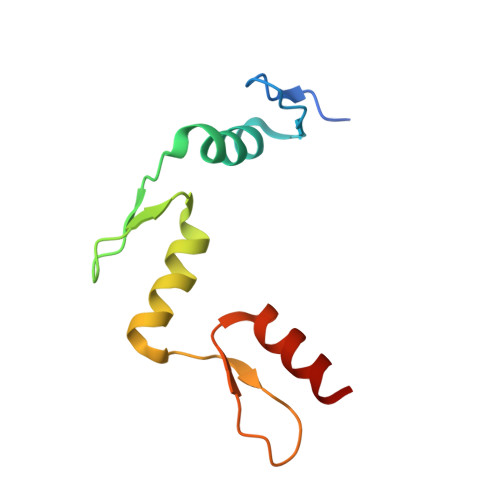
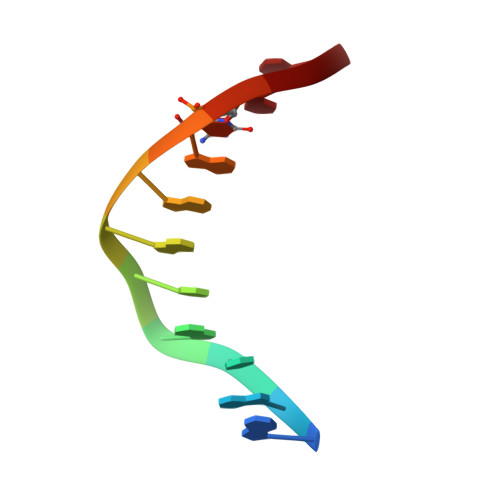
Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Hashimoto, H., Zhang, X., Zheng, Y., Wilson, G.G., Cheng, X.(2016) Nucleic Acids Res 44: 10165-10176
- PubMed: 27596598
- DOI: https://doi.org/10.1093/nar/gkw766
- Primary Citation of Related Structures:
5KL2, 5KL3, 5KL4, 5KL5, 5KL6, 5KL7 - PubMed Abstract:
Mutations in human zinc-finger transcription factor WT1 result in abnormal development of the kidneys and genitalia and an array of pediatric problems including nephropathy, blastoma, gonadal dysgenesis and genital discordance. Several overlapping phenotypes are associated with WT1 mutations, including Wilms tumors, Denys-Drash syndrome (DDS), Frasier syndrome (FS) and WAGR syndrome (Wilms tumor, aniridia, genitourinary malformations, and mental retardation). These conditions vary in severity from individual to individual; they can be fatal in early childhood, or relatively benign into adulthood. DDS mutations cluster predominantly in zinc fingers (ZF) 2 and 3 at the C-terminus of WT1, which together with ZF4 determine the sequence-specificity of DNA binding. We examined three DDS associated mutations in ZF2 of human WT1 where the normal glutamine at position 369 is replaced by arginine (Q369R), lysine (Q369K) or histidine (Q369H). These mutations alter the sequence-specificity of ZF2, we find, changing its affinity for certain bases and certain epigenetic forms of cytosine. X-ray crystallography of the DNA binding domains of normal WT1, Q369R and Q369H in complex with preferred sequences revealed the molecular interactions responsible for these affinity changes. DDS is inherited in an autosomal dominant fashion, implying a gain of function by mutant WT1 proteins. This gain, we speculate, might derive from the ability of the mutant proteins to sequester WT1 into unproductive oligomers, or to erroneously bind to variant target sequences.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
Organizational Affiliation: