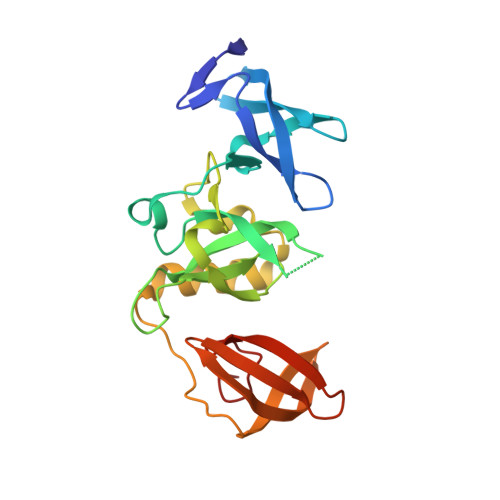
Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a
Iqbal, A., Mader, P., Dong, A., Dobrovetsky, E., Ferreira de Freitas, R., Walker, J.R., Bountra, C., Arrowsmith, C.H., Edwards, A.M., Schapira, M., Brown, P.J.To be published.