Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
Frey, B.F., Jiang, J., Sui, Y., Boyd, L.F., Yu, B., Tatsuno, G., Billeskov, R., Solaymani-Mohammadi, S., Berman, P.W., Margulies, D.H., Berzofsky, J.A.(2018) J Immunol 200: 1853-1864
- PubMed: 29374075
- DOI: https://doi.org/10.4049/jimmunol.1701523
- Primary Citation of Related Structures:
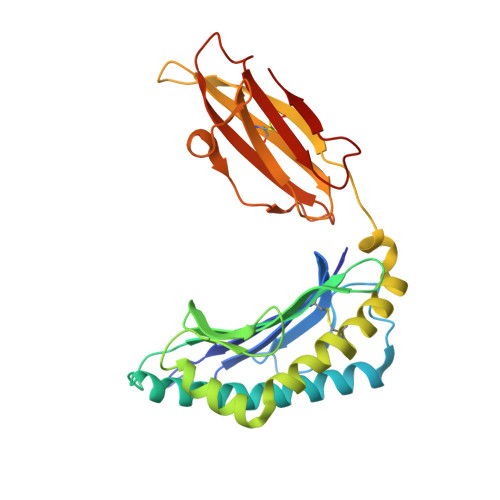
5KD4, 5KD7, 5T7G - PubMed Abstract:
Unlike cytosolic processing and presentation of viral Ags by virus-infected cells, Ags first expressed in infected nonprofessional APCs, such as CD4 + T cells in the case of HIV, are taken up by dendritic cells and cross-presented. This generally requires entry through the endocytic pathway, where endosomal proteases have first access for processing. Thus, understanding virus escape during cross-presentation requires an understanding of resistance to endosomal proteases, such as cathepsin S (CatS). We have modified HIV-1 MN gp120 by mutating a key CatS cleavage site (Thr 322 Thr 323 ) in the V3 loop of the immunodominant epitope IGPGRAFY TT to IGPGRAFY VV to prevent digestion. We found this mutation to facilitate cross-presentation and provide evidence from MHC binding and X-ray crystallographic structural studies that this results from preservation of the epitope rather than an increased epitope affinity for the MHC class I molecule. In contrast, when the protein is expressed by a vaccinia virus in the cytosol, the wild-type protein is immunogenic without this mutation. These proof-of-concept results show that a virus like HIV, infecting predominantly nonprofessional presenting cells, can escape T cell recognition by incorporating a CatS cleavage site that leads to destruction of an immunodominant epitope when the Ag undergoes endosomal cross-presentation.
- Vaccine Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892.
Organizational Affiliation: