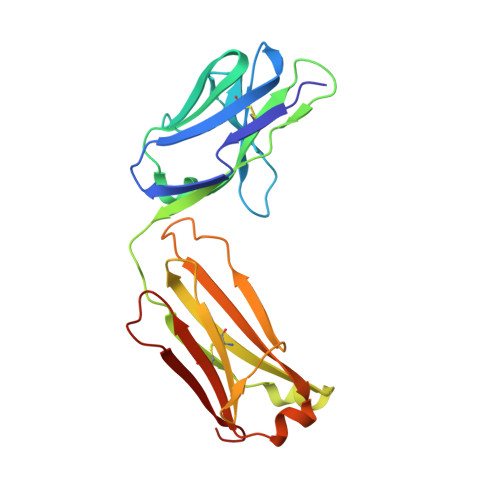
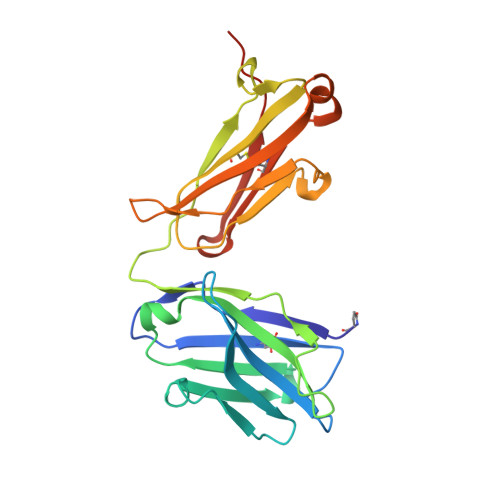
Data on crystal organization in the structure of the Fab fragment from the NIST reference antibody, RM 8671.
Gallagher, D.T., Karageorgos, I., Hudgens, J.W., Galvin, C.V.(2018) Data Brief 16: 29-36
- PubMed: 29167817
- DOI: https://doi.org/10.1016/j.dib.2017.11.013
- Primary Citation of Related Structures:
5K8A - PubMed Abstract:
The reported data describe the crystallization, crystal packing, structure determination and twinning of the unliganded Fab (antigen-binding fragment) from the NISTmAb (standard reference material 8671). The raw atomic coordinates are available as Protein Data Bank structure 5K8A and biological aspects are described in the article, (Karageorgos et al., 2017) [1]. Crystal data show that the packing is unique, and show the basis for the crystal's twinned growth. Twinning is a common and often serious problem in protein structure determination by x-ray crystallography [2]. In the present case the twinning is due to a small deviation (about 0.3 nm) from 4-fold symmetry in the primary intermolecular interface. The deviation produces pseudosymmetry, generating slightly different conformations of the protein, and alternating strong and weak forms of key packing interfaces throughout the lattice.
- Biomolecular Measurement Division, National Institute of Standards & Technology, Rockville, MD 20850, United States.
Organizational Affiliation: