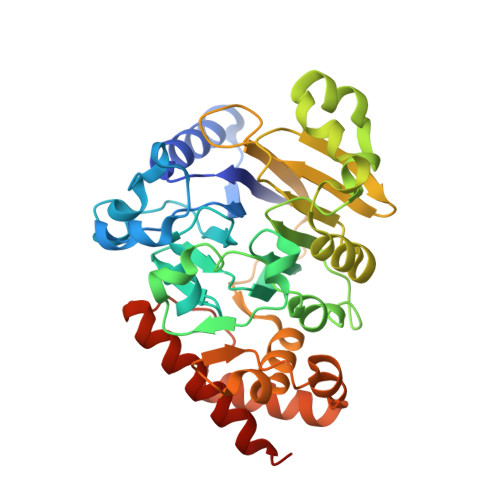
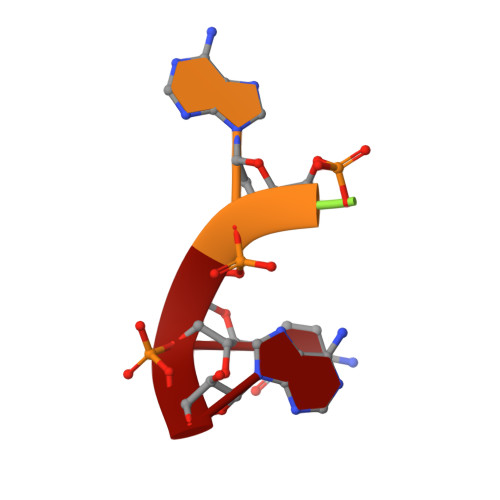
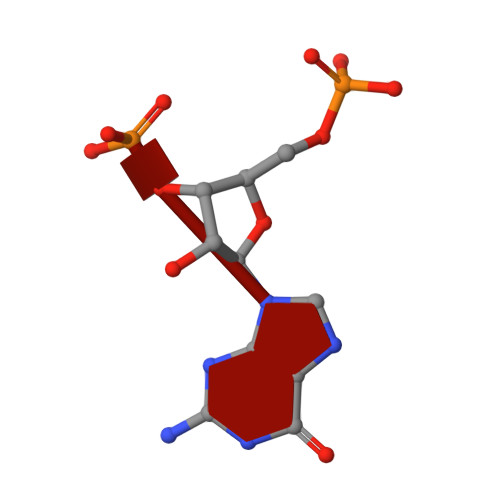
Metal dependence and branched RNA cocrystal structures of the RNA lariat debranching enzyme Dbr1.
Clark, N.E., Katolik, A., Roberts, K.M., Taylor, A.B., Holloway, S.P., Schuermann, J.P., Montemayor, E.J., Stevens, S.W., Fitzpatrick, P.F., Damha, M.J., Hart, P.J.(2016) Proc Natl Acad Sci U S A 113: 14727-14732
- PubMed: 27930312
- DOI: https://doi.org/10.1073/pnas.1612729114
- Primary Citation of Related Structures:
5K77 - PubMed Abstract:
Intron lariats are circular, branched RNAs (bRNAs) produced during pre-mRNA splicing. Their unusual chemical and topological properties arise from branch-point nucleotides harboring vicinal 2',5'- and 3',5'-phosphodiester linkages. The 2',5'-bonds must be hydrolyzed by the RNA debranching enzyme Dbr1 before spliced introns can be degraded or processed into small nucleolar RNA and microRNA derived from intronic RNA. Here, we measure the activity of Dbr1 from Entamoeba histolytica by using a synthetic, dark-quenched bRNA substrate that fluoresces upon hydrolysis. Purified enzyme contains nearly stoichiometric equivalents of Fe and Zn per polypeptide and demonstrates turnover rates of ∼3 s -1 Similar rates are observed when apo-Dbr1 is reconstituted with Fe(II)+Zn(II) under aerobic conditions. Under anaerobic conditions, a rate of ∼4.0 s -1 is observed when apoenzyme is reconstituted with Fe(II). In contrast, apo-Dbr1 reconstituted with Mn(II) or Fe(II) under aerobic conditions is inactive. Diffraction data from crystals of purified enzyme using X-rays tuned to the Fe absorption edge show Fe partitions primarily to the β-pocket and Zn to the α-pocket. Structures of the catalytic mutant H91A in complex with 7-mer and 16-mer synthetic bRNAs reveal bona fide RNA branchpoints in the Dbr1 active site. A bridging hydroxide is in optimal position for nucleophilic attack of the scissile phosphate. The results clarify uncertainties regarding structure/function relationships in Dbr1 enzymes, and the fluorogenic probe permits high-throughput screening for inhibitors that may hold promise as treatments for retroviral infections and neurodegenerative disease.
- Department of Biochemistry and Structural Biology, University of Texas Health Science Center, San Antonio, TX 78229; clarkn@uthscsa.edu fitzpatrickp@uthscsa.edu masad.damha@mcgill.ca pjh@biochem.uthscsa.edu.
Organizational Affiliation: