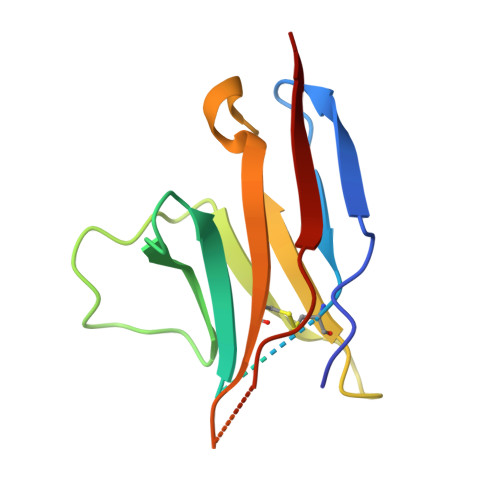
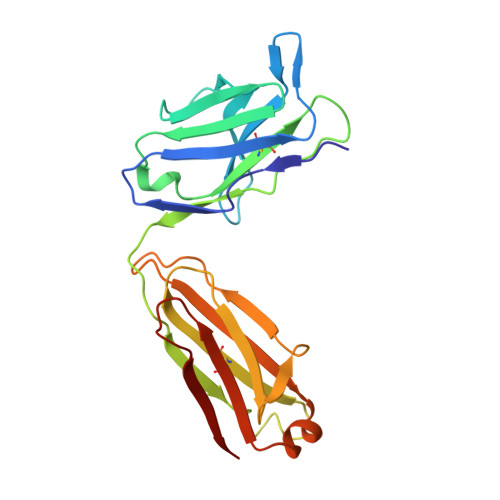
Structural basis for blocking PD-1-mediated immune suppression by therapeutic antibody pembrolizumab.
Na, Z.,
Yeo, S.P.,
Bharath, S.R.,
Bowler, M.W.,
Balijkcij, E.,
Wang, C.I.,
Song, H.(2017) Cell Res 27: 147-150
- PubMed: 27325296 Search on PubMedSearch on PubMed Central
- DOI: https://doi.org/10.1038/cr.2016.77
- Primary Citation of Related Structures:
5JXE
Organizational Affiliation: - Institute of Molecular and Cell Biology, 61 Biopolis Drive, Singapore 138673, Singapore.
- Singapore Immunology Network, 8a Biomedical Grove, Singapore 138648, Singapore.
- European Molecular Biology Laboratory, Grenoble Outstation, 71 avenue des Martyrs, CS 90181 F-38042 Grenoble, France.
- Unit of Virus Host-Cell Interactions, Univ. Grenoble Alpes-EMBL-CNRS, 71 avenue des Martyrs, CS 90181 F-38042 Grenoble, France.
- Department of Biochemistry, National University of Singapore, 14 Science Drive, Singapore 117543, Singapore.