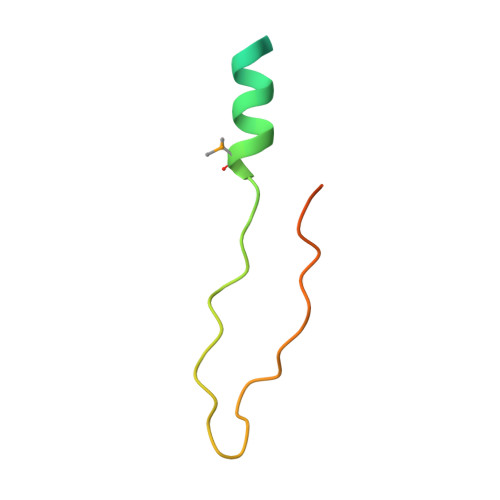
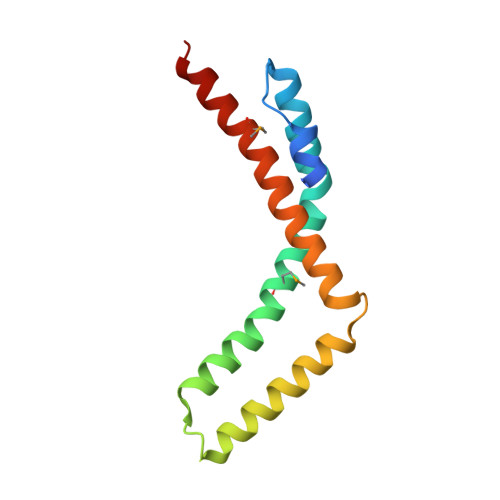
Structural basis for ELL2 and AFF4 activation of HIV-1 proviral transcription.
Qi, S., Li, Z., Schulze-Gahmen, U., Stjepanovic, G., Zhou, Q., Hurley, J.H.(2017) Nat Commun 8: 14076-14076
- PubMed: 28134250
- DOI: https://doi.org/10.1038/ncomms14076
- Primary Citation of Related Structures:
5JW9 - PubMed Abstract:
The intrinsically disordered scaffold proteins AFF1/4 and the transcription elongation factors ELL1/2 are core components of the super elongation complex required for HIV-1 proviral transcription. Here we report the 2.0-Å resolution crystal structure of the human ELL2 C-terminal domain bound to its 50-residue binding site on AFF4, the ELLBow. The ELL2 domain has the same arch-shaped fold as the tight junction protein occludin. The ELLBow consists of an N-terminal helix followed by an extended hairpin that we refer to as the elbow joint, and occupies most of the concave surface of ELL2. This surface is important for the ability of ELL2 to promote HIV-1 Tat-mediated proviral transcription. The AFF4-ELL2 interface is imperfectly packed, leaving a cavity suggestive of a potential binding site for transcription-promoting small molecules.
- Department of Urology, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University and National Collaborative Innovation Center, Chengdu 610041, China.
Organizational Affiliation: