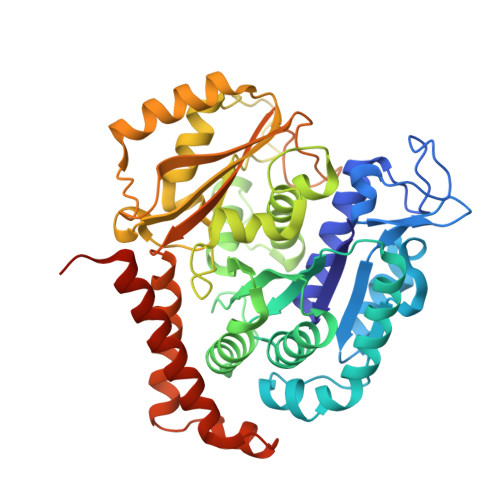
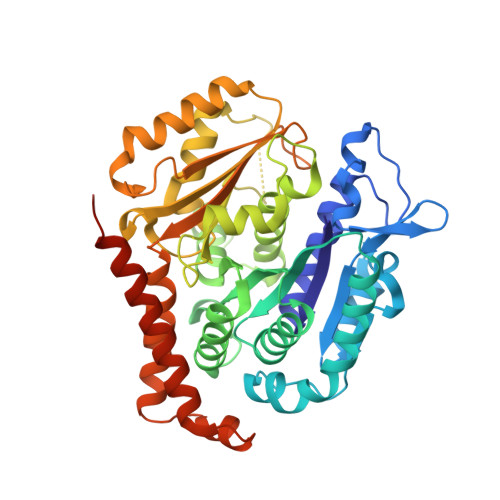
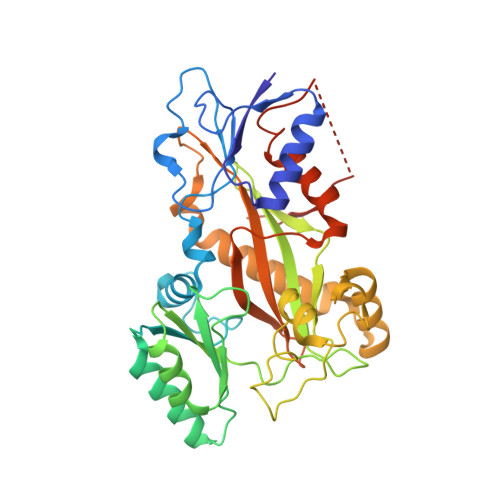
Antivascular and antitumor properties of the tubulin-binding chalcone TUB091.
Canela, M.D., Noppen, S., Bueno, O., Prota, A.E., Bargsten, K., Saez-Calvo, G., Jimeno, M.L., Benkheil, M., Ribatti, D., Velazquez, S., Camarasa, M.J., Diaz, J.F., Steinmetz, M.O., Priego, E.M., Perez-Perez, M.J., Liekens, S.(2017) Oncotarget 8: 14325-14342
- PubMed: 27224920
- DOI: https://doi.org/10.18632/oncotarget.9527
- Primary Citation of Related Structures:
5JVD - PubMed Abstract:
We investigated the microtubule-destabilizing, vascular-targeting, anti-tumor and anti-metastatic activities of a new series of chalcones, whose prototype compound is (E)-3-(3''-amino-4''-methoxyphenyl)-1-(5'-methoxy-3',4'-methylendioxyphenyl)-2-methylprop-2-en-1-one (TUB091). X-ray crystallography showed that these chalcones bind to the colchicine site of tubulin and therefore prevent the curved-to-straight structural transition of tubulin, which is required for microtubule formation. Accordingly, TUB091 inhibited cancer and endothelial cell growth, induced G2/M phase arrest and apoptosis at 1-10 nM. In addition, TUB091 displayed vascular disrupting effects in vitro and in the chicken chorioallantoic membrane (CAM) assay at low nanomolar concentrations. A water-soluble L-Lys-L-Pro derivative of TUB091 (i.e. TUB099) showed potent antitumor activity in melanoma and breast cancer xenograft models by causing rapid intratumoral vascular shutdown and massive tumor necrosis. TUB099 also displayed anti-metastatic activity similar to that of combretastatin A4-phosphate. Our data indicate that this novel class of chalcones represents interesting lead molecules for the design of vascular disrupting agents (VDAs). Moreover, we provide evidence that our prodrug approach may be valuable for the development of anti-cancer drugs.
- Instituto de Química Médica (IQM-CSIC), Madrid, Spain.
Organizational Affiliation: