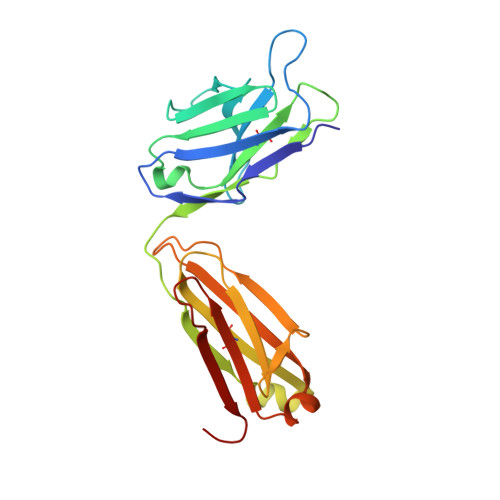
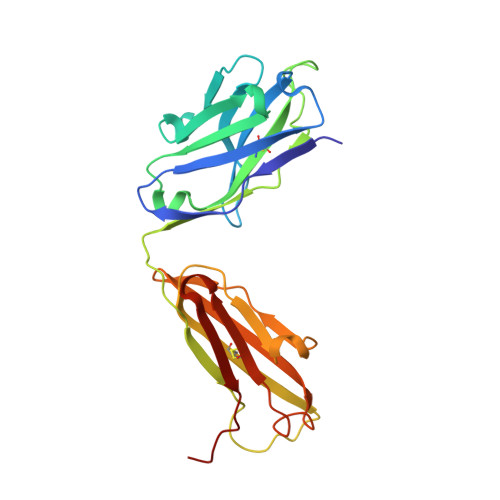
Crystal structure of the antigen-binding fragment of a monoclonal antibody specific for the multidrug-resistance-linked ABC transporter human P-glycoprotein.
Esser, L., Shukla, S., Zhou, F., Ambudkar, S.V., Xia, D.(2016) Acta Crystallogr F Struct Biol Commun 72: 636-641
- PubMed: 27487928
- DOI: https://doi.org/10.1107/S2053230X16009778
- Primary Citation of Related Structures:
5JUE - PubMed Abstract:
P-glycoprotein (P-gp) is a polyspecific ATP-dependent transporter linked to multidrug resistance in cancers that plays important roles in the pharmacokinetics of a large number of drugs. The drug-resistance phenotype of P-gp can be modulated by the monoclonal antibody UIC2, which specifically recognizes human P-gp in a conformation-dependent manner. Here, the purification, sequence determination and high-resolution structure of the Fab fragment of UIC2 (UIC2/Fab) are reported. Purified UIC2/Fab binds human P-gp with a 1:1 stoichiometry. Crystals of UIC2/Fab are triclinic (space group P1), with unit-cell parameters a = 40.67, b = 44.91, c = 58.09 Å, α = 97.62, β = 99.10, γ = 94.09°, and diffracted X-rays to 1.6 Å resolution. The structure was determined by molecular replacement and refined to 1.65 Å resolution. The asymmetric unit contains one molecule of UIC2/Fab, which exhibits a positively charged antigen-binding surface, suggesting that it might recognize an oppositely charged extracellular epitope of P-gp.
- National Cancer Institute, National Institutes of Health, 37 Convent Drive, Building 37, Bethesda, MD 20892, USA.
Organizational Affiliation: