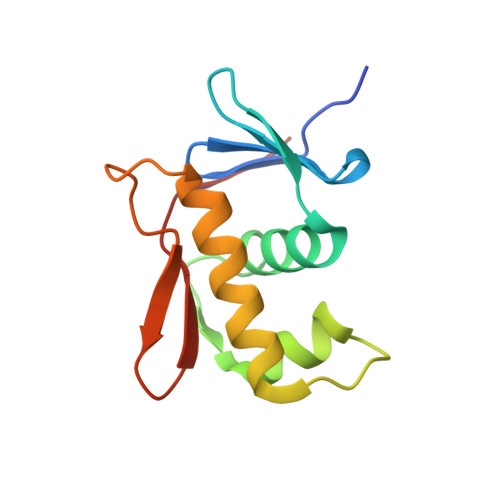
Structure-function analysis of the DNA-binding domain of a transmembrane transcriptional activator.
Schlundt, A., Buchner, S., Janowski, R., Heydenreich, T., Heermann, R., Lassak, J., Geerlof, A., Stehle, R., Niessing, D., Jung, K., Sattler, M.(2017) Sci Rep 7: 1051-1051
- PubMed: 28432336
- DOI: https://doi.org/10.1038/s41598-017-01031-9
- Primary Citation of Related Structures:
5JU7 - PubMed Abstract:
The transmembrane DNA-binding protein CadC of E. coli, a representative of the ToxR-like receptor family, combines input and effector domains for signal sensing and transcriptional activation, respectively, in a single protein, thus representing one of the simplest signalling systems. At acidic pH in a lysine-rich environment, CadC activates the transcription of the cadBA operon through recruitment of the RNA polymerase (RNAP) to the two cadBA promoter sites, Cad1 and Cad2, which are directly bound by CadC. However, the molecular details for its interaction with DNA have remained elusive. Here, we present the crystal structure of the CadC DNA-binding domain (DBD) and show that it adopts a winged helix-turn-helix fold. The interaction with the cadBA promoter site Cad1 is studied by using nuclear magnetic resonance (NMR) spectroscopy, biophysical methods and functional assays and reveals a preference for AT-rich regions. By mutational analysis we identify amino acids within the CadC DBD that are crucial for DNA-binding and functional activity. Experimentally derived structural models of the CadC-DNA complex indicate that the CadC DBD employs mainly non-sequence-specific over a few specific contacts. Our data provide molecular insights into the CadC-DNA interaction and suggest how CadC dimerization may provide high-affinity binding to the Cad1 promoter.
- Munich Center for Integrated Protein Science (CiPSM) at the Department of Chemistry, Technische Universität München, 85748, Garching, Germany.
Organizational Affiliation: