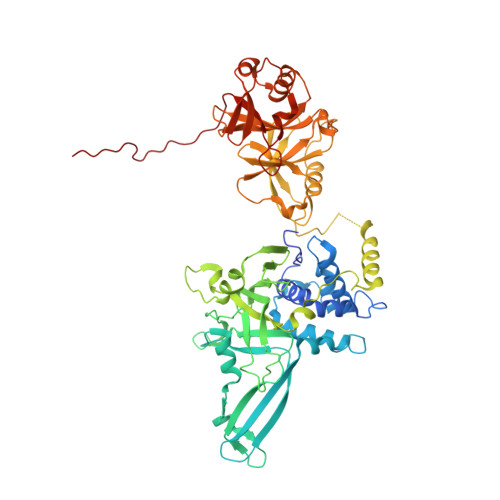
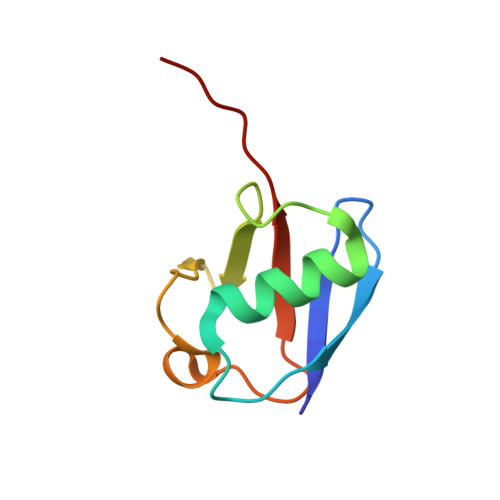
Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation.
Rouge, L., Bainbridge, T.W., Kwok, M., Tong, R., Di Lello, P., Wertz, I.E., Maurer, T., Ernst, J.A., Murray, J.(2016) Structure 24: 1335-1345
- PubMed: 27452404
- DOI: https://doi.org/10.1016/j.str.2016.05.020
- Primary Citation of Related Structures:
5J7T, 5JTJ, 5JTV - PubMed Abstract:
The deubiquitinating enzyme USP7 has a pivotal role in regulating the stability of proteins involved in fundamental cellular processes of normal biology and disease. Despite the importance of USP7, the mechanisms underlying substrate recognition and catalytic activation are poorly understood. Here we present structural, biochemical, and biophysical analyses elucidating the molecular mechanism by which the C-terminal 19 amino acids of USP7 (residues 1084-1102) enhance the ubiquitin cleavage activity of the deubiquitinase (DUB) domain. Our data demonstrate that the C-terminal peptide binds the activation cleft in the catalytic domain and stabilizes the catalytically competent conformation of USP7. Additional structures of longer fragments of USP7, as well as solution studies, provide insight into full-length USP7, the role of the UBL domains, and demonstrate that both substrate recognition and deubiquitinase activity are highly regulated by the catalytic and noncatalytic domains of USP7, a feature that could be essential for the proper function of multi-domain DUBs.
- Department of Structural Biology, Genentech, South San Francisco, CA 94080, USA.
Organizational Affiliation: