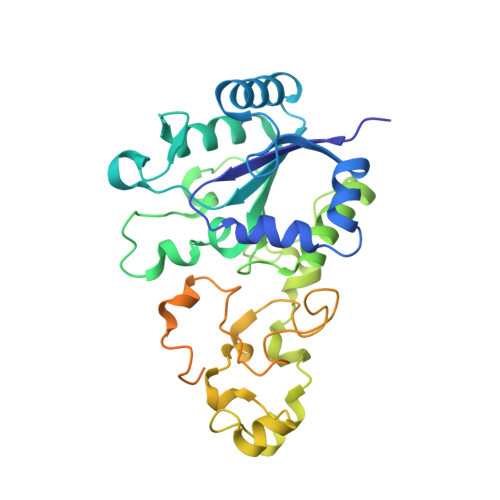
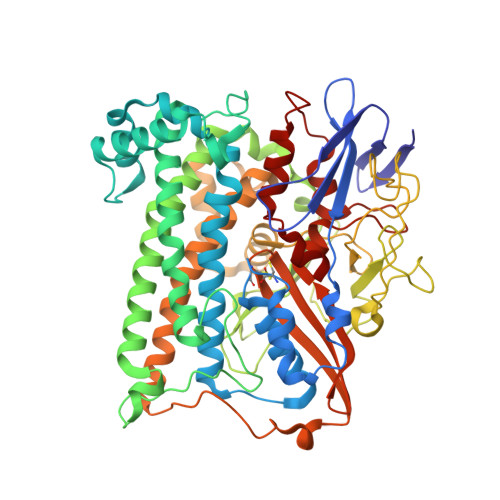
Importance of the Active Site "Canopy" Residues in an O2-Tolerant [NiFe]-Hydrogenase.
Brooke, E.J., Evans, R.M., Islam, S.T., Roberts, G.M., Wehlin, S.A., Carr, S.B., Phillips, S.E., Armstrong, F.A.(2017) Biochemistry 56: 132-142
- PubMed: 28001048
- DOI: https://doi.org/10.1021/acs.biochem.6b00868
- Primary Citation of Related Structures:
5JRD - PubMed Abstract:
The active site of Hyd-1, an oxygen-tolerant membrane-bound [NiFe]-hydrogenase from Escherichia coli, contains four highly conserved residues that form a "canopy" above the bimetallic center, closest to the site at which exogenous agents CO and O 2 interact, substrate H 2 binds, and a hydrido intermediate is stabilized. Genetic modification of the Hyd-1 canopy has allowed the first systematic and detailed kinetic and structural investigation of the influence of the immediate outer coordination shell on H 2 activation. The central canopy residue, arginine 509, suspends a guanidine/guanidinium side chain at close range above the open coordination site lying between the Ni and Fe atoms (N-metal distance of 4.4 Å): its replacement with lysine lowers the H 2 oxidation rate by nearly 2 orders of magnitude and markedly decreases the H 2 /D 2 kinetic isotope effect. Importantly, this collapse in rate constant can now be ascribed to a very unfavorable activation entropy (easily overriding the more favorable activation enthalpy of the R509K variant). The second most important canopy residue for H 2 oxidation is aspartate 118, which forms a salt bridge to the arginine 509 headgroup: its mutation to alanine greatly decreases the H 2 oxidation efficiency, observed as a 10-fold increase in the potential-dependent Michaelis constant. Mutations of aspartate 574 (also salt-bridged to R509) to asparagine and proline 508 to alanine have much smaller effects on kinetic properties. None of the mutations significantly increase sensitivity to CO, but neutralizing the expected negative charges from D118 and D574 decreases O 2 tolerance by stabilizing the oxidized resting Ni III -OH state ("Ni-B"). An extensive model of the catalytic importance of residues close to the active site now emerges, whereby a conserved gas channel culminates in the arginine headgroup suspended above the Ni and Fe.
- Department of Chemistry, University of Oxford , Oxford, U.K.
Organizational Affiliation: