Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Lu, J., Chan, C., Yu, L., Fan, J., Sun, F., Zhai, Y.(2020) Commun Biol
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein UPS1, mitochondrial,Mitochondrial distribution and morphology protein 35 | 283 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: UPS1, YLR193C, MDM35, YKL053C-A Membrane Entity: Yes | 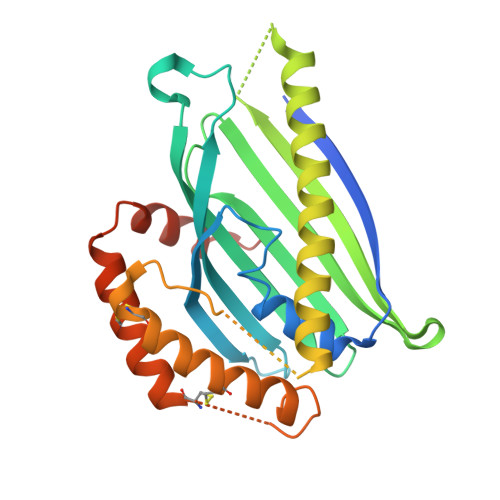 | |
UniProt | |||||
Find proteins for O60200 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore O60200 Go to UniProtKB: O60200 | |||||
Find proteins for Q05776 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q05776 Go to UniProtKB: Q05776 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | O60200Q05776 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 118.563 | α = 90 |
| b = 68.29 | β = 102.1 |
| c = 105.137 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data scaling |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data processing |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| China | -- |