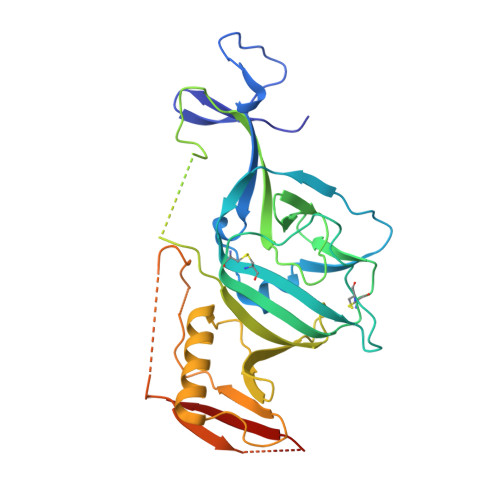
Toremifene interacts with and destabilizes the Ebola virus glycoprotein.
Zhao, Y., Ren, J., Harlos, K., Jones, D.M., Zeltina, A., Bowden, T.A., Padilla-Parra, S., Fry, E.E., Stuart, D.I.(2016) Nature 535: 168-172
- PubMed: 27362232
- DOI: https://doi.org/10.1038/nature18615
- Primary Citation of Related Structures:
5JQ3, 5JQ7, 5JQB - PubMed Abstract:
Ebola viruses (EBOVs) are responsible for repeated outbreaks of fatal infections, including the recent deadly epidemic in West Africa. There are currently no approved therapeutic drugs or vaccines for the disease. EBOV has a membrane envelope decorated by trimers of a glycoprotein (GP, cleaved by furin to form GP1 and GP2 subunits), which is solely responsible for host cell attachment, endosomal entry and membrane fusion. GP is thus a primary target for the development of antiviral drugs. Here we report the first, to our knowledge, unliganded structure of EBOV GP, and high-resolution complexes of GP with the anticancer drug toremifene and the painkiller ibuprofen. The high-resolution apo structure gives a more complete and accurate picture of the molecule, and allows conformational changes introduced by antibody and receptor binding to be deciphered. Unexpectedly, both toremifene and ibuprofen bind in a cavity between the attachment (GP1) and fusion (GP2) subunits at the entrance to a large tunnel that links with equivalent tunnels from the other monomers of the trimer at the three-fold axis. Protein–drug interactions with both GP1 and GP2 are predominately hydrophobic. Residues lining the binding site are highly conserved among filoviruses except Marburg virus (MARV), suggesting that MARV may not bind these drugs. Thermal shift assays show up to a 14 °C decrease in the protein melting temperature after toremifene binding, while ibuprofen has only a marginal effect and is a less potent inhibitor. These results suggest that inhibitor binding destabilizes GP and triggers premature release of GP2, thereby preventing fusion between the viral and endosome membranes. Thus, these complex structures reveal the mechanism of inhibition and may guide the development of more powerful anti-EBOV drugs.
- Division of Structural Biology, University of Oxford, The Henry Wellcome Building for Genomic Medicine, Headington, Oxford, OX3 7BN, UK.
Organizational Affiliation: