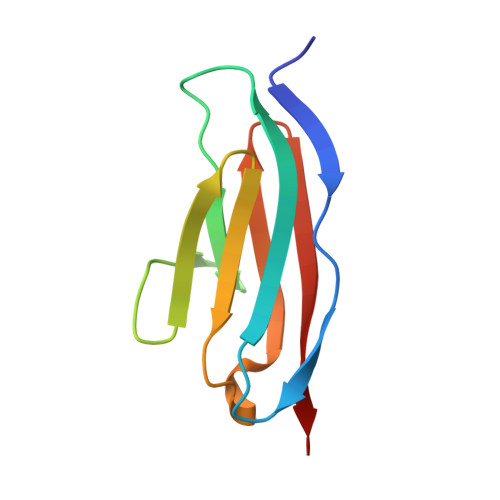
CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex.
Zhou, T., Fleming, J.R., Franke, B., Bogomolovas, J., Barsukov, I., Rigden, D.J., Labeit, S., Mayans, O.(2016) FEBS Lett 590: 3098-3110
- PubMed: 27531639
- DOI: https://doi.org/10.1002/1873-3468.12362
- Primary Citation of Related Structures:
5JOE - PubMed Abstract:
The cardiac ankyrin repeat protein (CARP) is up-regulated in the myocardium during cardiovascular disease and in response to mechanical or toxic stress. Stress-induced CARP interacts with the N2A spring region of the titin filament to modulate muscle compliance. We characterize the interaction between CARP and titin-N2A and show that the binding site in titin spans the dual domain UN2A-Ig81. We find that the unique sequence UN2A is not structurally disordered, but that it has a stable, elongated α-helical fold that possibly acts as a constant force spring. Our findings portray CARP/titin-N2A as a structured node and help to rationalize the molecular basis of CARP mechanosensing in the sarcomeric I-band.
- Department of Biology, University of Konstanz, Germany.
Organizational Affiliation: