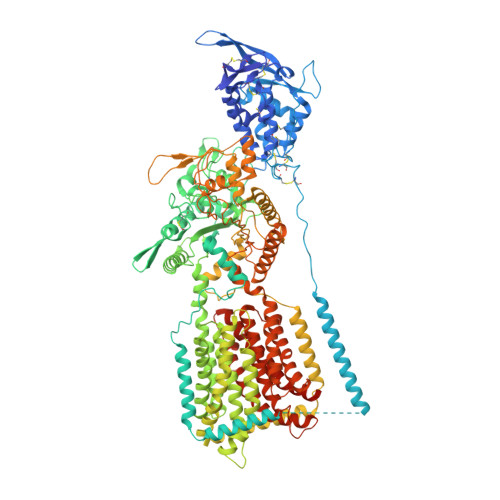
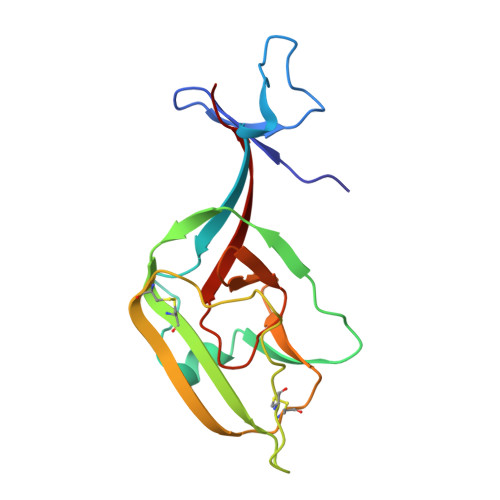
Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection
Gong, X., Qian, H., Zhou, X., Wu, J., Wan, T., Cao, P., Huang, W., Zhao, X., Wang, X., Wang, P., Shi, Y., Gao, G.F., Zhou, Q., Yan, N.(2016) Cell 165: 1467-1478
- PubMed: 27238017
- DOI: https://doi.org/10.1016/j.cell.2016.05.022
- Primary Citation of Related Structures:
5JNX - PubMed Abstract:
Niemann-Pick disease type C (NPC) is associated with mutations in NPC1 and NPC2, whose gene products are key players in the endosomal/lysosomal egress of low-density lipoprotein-derived cholesterol. NPC1 is also the intracellular receptor for Ebola virus (EBOV). Here, we present a 4.4 Å structure of full-length human NPC1 and a low-resolution reconstruction of NPC1 in complex with the cleaved glycoprotein (GPcl) of EBOV, both determined by single-particle electron cryomicroscopy. NPC1 contains 13 transmembrane segments (TMs) and three distinct lumenal domains A (also designated NTD), C, and I. TMs 2-13 exhibit a typical resistance-nodulation-cell division fold, among which TMs 3-7 constitute the sterol-sensing domain conserved in several proteins involved in cholesterol metabolism and signaling. A trimeric EBOV-GPcl binds to one NPC1 monomer through the domain C. Our structural and biochemical characterizations provide an important framework for mechanistic understanding of NPC1-mediated intracellular cholesterol trafficking and Ebola virus infection.
- State Key Laboratory of Membrane Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China; Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China; Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: