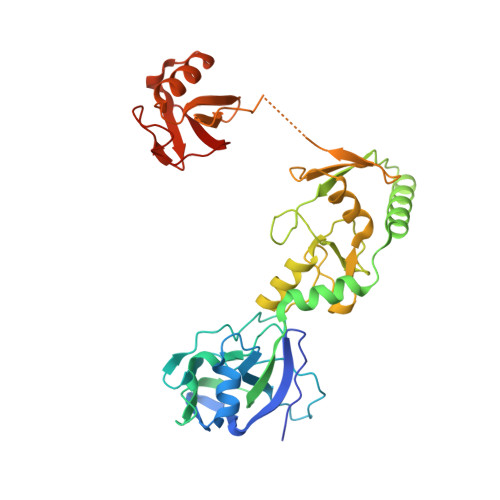
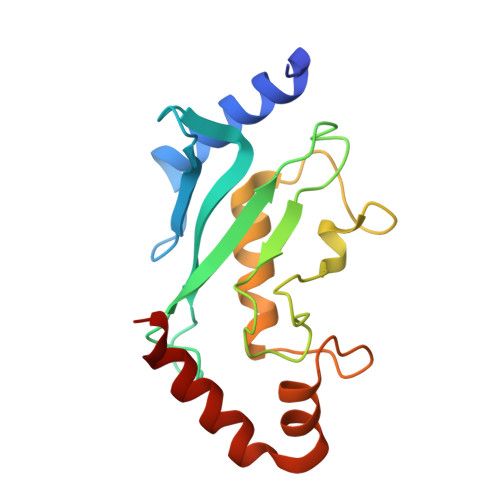
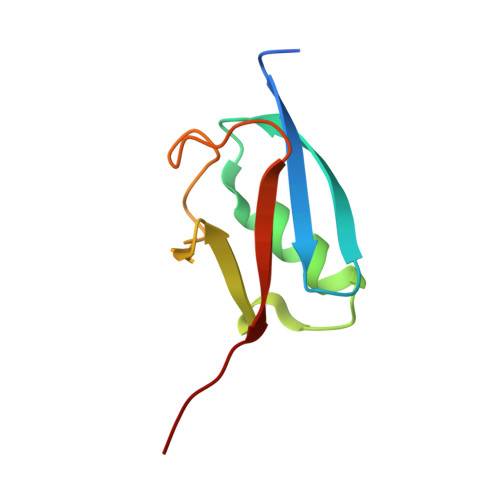
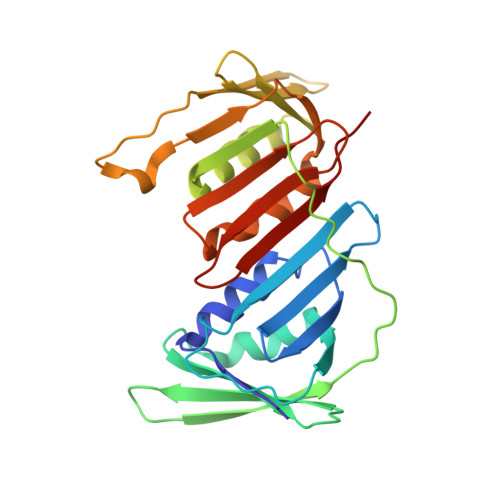
Capturing a substrate in an activated RING E3/E2-SUMO complex.
Streich, F.C., Lima, C.D.(2016) Nature 536: 304-308
- PubMed: 27509863
- DOI: https://doi.org/10.1038/nature19071
- Primary Citation of Related Structures:
5JNE - PubMed Abstract:
Post-translational protein modification by ubiquitin (Ub) and ubiquitin-like (Ubl) proteins such as small ubiquitin-like modifier (SUMO) regulates processes including protein homeostasis, the DNA damage response, and the cell cycle. Proliferating cell nuclear antigen (PCNA) is modified by Ub or poly-Ub at lysine (Lys)164 after DNA damage to recruit repair factors. Yeast PCNA is modified by SUMO on Lys164 and Lys127 during S-phase to recruit the anti-recombinogenic helicase Srs2. Lys164 modification requires specialized E2/E3 enzyme pairs for SUMO or Ub conjugation. For SUMO, Lys164 modification is strictly dependent on the E3 ligase Siz1, suggesting the E3 alters E2 specificity to promote Lys164 modification. The structural basis for substrate interactions in activated E3/E2–Ub/Ubl complexes remains unclear. Here we report an engineered E2 protein and cross-linking strategies that trap an E3/E2–Ubl/substrate complex for structure determination, illustrating how an E3 can bypass E2 specificity to force-feed a substrate lysine into the E2 active site.